Predicting Therapeutic and Aggravating Drugs for Hepatocellular Carcinoma Based on Tissue- Specific Pathways
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Microneedle Device and Transdermal Administration Device Provided with Microneedles
(19) & (11) EP 2 005 990 A9 (12) CORRECTED EUROPEAN PATENT APPLICATION published in accordance with Art. 153(4) EPC (15) Correction information: (51) Int Cl.: Corrected version no 1 (W1 A1) A61M 37/00 (2006.01) A61B 17/20 (2006.01) Corrections, see A61K 9/00 (2006.01) A61K 47/34 (2006.01) Search report Search Report replaced or added (86) International application number: PCT/JP2007/057737 (48) Corrigendum issued on: 29.07.2009 Bulletin 2009/31 (87) International publication number: WO 2007/116959 (18.10.2007 Gazette 2007/42) (43) Date of publication: 24.12.2008 Bulletin 2008/52 (21) Application number: 07741173.4 (22) Date of filing: 06.04.2007 (84) Designated Contracting States: • MATSUDO, Toshiyuki AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Tsukuba-shi Ibaraki 305-0856 (JP) HU IE IS IT LI LT LU LV MC MT NL PL PT RO SE • KUWAHARA, Tetsuji SI SK TR Tsukuba-shi Ibaraki 305-0856 (JP) (30) Priority: 07.04.2006 JP 2006106995 (74) Representative: Westendorp, Michael Oliver Splanemann Reitzner (71) Applicant: Hisamitsu Pharmaceutical Co., Inc. Baronetzky Westendorp Tosu-shi, Saga 841 (JP) Rumfordstrasse 7 D-80469 München (DE) (72) Inventors: • TOKUMOTO, Seiji Tsukuba-shi Ibaraki 305-0856 (JP) (54) MICRONEEDLE DEVICE AND TRANSDERMAL ADMINISTRATION DEVICE PROVIDED WITH MICRONEEDLES (57) The present invention provides a microneedle device having a coating, which is effective even with a low molecular weight active compound and can sustain the effect of the drug for a long period of time, and a transdermal drug administration apparatus with micro- needles. -

The In¯Uence of Medication on Erectile Function
International Journal of Impotence Research (1997) 9, 17±26 ß 1997 Stockton Press All rights reserved 0955-9930/97 $12.00 The in¯uence of medication on erectile function W Meinhardt1, RF Kropman2, P Vermeij3, AAB Lycklama aÁ Nijeholt4 and J Zwartendijk4 1Department of Urology, Netherlands Cancer Institute/Antoni van Leeuwenhoek Hospital, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands; 2Department of Urology, Leyenburg Hospital, Leyweg 275, 2545 CH The Hague, The Netherlands; 3Pharmacy; and 4Department of Urology, Leiden University Hospital, P.O. Box 9600, 2300 RC Leiden, The Netherlands Keywords: impotence; side-effect; antipsychotic; antihypertensive; physiology; erectile function Introduction stopped their antihypertensive treatment over a ®ve year period, because of side-effects on sexual function.5 In the drug registration procedures sexual Several physiological mechanisms are involved in function is not a major issue. This means that erectile function. A negative in¯uence of prescrip- knowledge of the problem is mainly dependent on tion-drugs on these mechanisms will not always case reports and the lists from side effect registries.6±8 come to the attention of the clinician, whereas a Another way of looking at the problem is drug causing priapism will rarely escape the atten- combining available data on mechanisms of action tion. of drugs with the knowledge of the physiological When erectile function is in¯uenced in a negative mechanisms involved in erectile function. The way compensation may occur. For example, age- advantage of this approach is that remedies may related penile sensory disorders may be compen- evolve from it. sated for by extra stimulation.1 Diminished in¯ux of In this paper we will discuss the subject in the blood will lead to a slower onset of the erection, but following order: may be accepted. -

Affinity Profiles of Hexahydro-Sila-Difenidol Analogues at Muscarinic Receptor Subtypes
European Journal of Pharmacology, 168 (1989) 71-80 71 Elsevier EJP 50940 Affinity profiles of hexahydro-sila-difenidol analogues at muscarinic receptor subtypes 1 Günter Lambrecht *,Roland Feifel, Monika Wagner-Röder, Carsten Strohmann , 1 1 2 2 Harald Zilch , Reinhold Tacke , Magali Waelbroeck , Jean Christophe , Hendrikus Boddeke 3 and Ernst Mutschier Department of Pharmacology, University of Frankfurt, D-6000 Frankfurt/ M, F.R.G., 1 Institute of Jnorganic Chemistry, University of Karlsruhe, D-7500 Karlsruhe, F.R.G., 1 Department of Biochemistry and Nutrition, Medica/ Schoo/, Free University of Brussels, B-1000 Brussels, Belgium, and 1 Preclinical Research, Sandoz Ltd., CH-4002 Basel, Switzerland Received 20 Apri11989, accepted 13 June 1989 In an attempt to assess the structural requirements of hexahydro-sila-difenidol for potency and selectivity, a series of analogues modified in the amino group and the phenyl ring were investigated for their affinity to muscarinic M1- (rabbit vas deferens), Mr (guinea-pig atria) and Mr (guinea-pig ileum) receptors. All compounds were competitive antagonists in the three tissues. Their affinities to the three muscarinic receptor subtypes differed by more than two orders of magnitude and the observed receptor selectivities were not associated with high affinity. The pyrrolidino and hexamethyleneimino analogues, compounds substituted in the phenylring with a methoxy group or a chlorine atom as weil as p-fluoro-hexahydro-difenidol displayed the same affinity profile as the parent compound, hexahydro-sila-difen idol: M1 =M 3 > M 2 • A different selectivity patternwas observed for p-fluoro-hexahydro-sila-difenidol: M3 > M1 > M 2 • This compound exhibited its highest affinity for M3-receptors in guinea-pig ileum (pA 2 = 7.84), intermediate affinity for M1-receptors in rabbit vas deferens (pA 2 = 6.68) and lowest affinity for the Mrreceptors in guinea-pig atria (pA 2 = 6.01). -

Pharmacokinetics of Oral Pridinol: Results of a Randomized, Crossover Bioequivalence Trial in Healthy Subjects
International Journal of Clinical Pharmacology and Therapeutics, Vol. 59 – No. 6/2021 (471-477) Pharmacokinetics of oral pridinol: Results of a randomized, crossover bioequivalence trial in healthy subjects Original Maren Richter1, Frank Donath1, Ralph-Steven Wedemeyer2, André Warnke1, 3 3 ©2021 Dustri-Verlag Dr. K. Feistle Andreas Horstmann , and Claudia Peschel ISSN 0946-1965 DOI 10.5414/CP203900 1SocraTec R&D GmbH, Oberursel/Erfurt, 2SocraMetrics GmbH, Erfurt, and e-pub: April 9, 2021 3Strathmann GmbH & Co. KG, Hamburg, Germany Key words Abstract. Objectives: To establish the What is known about this subject bioavailability – bio- relative bioavailability and to assess bio- equivalence – immedi- equivalence of oral, immediate-release tab- – The anticholinergic agent pridinol has ate-release tablet – lets containing pridinol and to determine been used as a muscle relaxant for de- pharmacokinetics the pharmacokinetic properties of the com- cades. – pridinol pound. Methods and materials: In this single- – However, the published literature on the center, open-label, randomized, crossover pharmacology of pridinol is sparse. trial, healthy male and female adult subjects received single doses of the test and refer- – More accessible information on the cha- ence product containing 4 mg pridinol me- racteristics of pridinol, e.g., its pharma- sylate (equivalent to 3 mg pridinol) each cokinetics in humans, is needed. under fasting conditions. For pharmacoki- netic evaluation, blood samples were with- What this study adds drawn until 72 hours post dose. Pridinol in plasma was quantified by validated liquid – This is the first detailed report on the chromatography-mass spectrometry/mass pharmacokinetic properties of pridinol in spectrometry (LC-MS/MS). Adverse events humans, assisting the prescriber to make (AEs) were analyzed descriptively. -
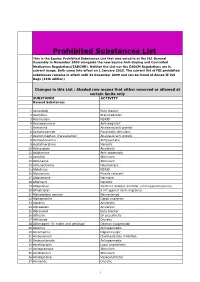
Prohibited Substances List
Prohibited Substances List This is the Equine Prohibited Substances List that was voted in at the FEI General Assembly in November 2009 alongside the new Equine Anti-Doping and Controlled Medication Regulations(EADCMR). Neither the List nor the EADCM Regulations are in current usage. Both come into effect on 1 January 2010. The current list of FEI prohibited substances remains in effect until 31 December 2009 and can be found at Annex II Vet Regs (11th edition) Changes in this List : Shaded row means that either removed or allowed at certain limits only SUBSTANCE ACTIVITY Banned Substances 1 Acebutolol Beta blocker 2 Acefylline Bronchodilator 3 Acemetacin NSAID 4 Acenocoumarol Anticoagulant 5 Acetanilid Analgesic/anti-pyretic 6 Acetohexamide Pancreatic stimulant 7 Acetominophen (Paracetamol) Analgesic/anti-pyretic 8 Acetophenazine Antipsychotic 9 Acetylmorphine Narcotic 10 Adinazolam Anxiolytic 11 Adiphenine Anti-spasmodic 12 Adrafinil Stimulant 13 Adrenaline Stimulant 14 Adrenochrome Haemostatic 15 Alclofenac NSAID 16 Alcuronium Muscle relaxant 17 Aldosterone Hormone 18 Alfentanil Narcotic 19 Allopurinol Xanthine oxidase inhibitor (anti-hyperuricaemia) 20 Almotriptan 5 HT agonist (anti-migraine) 21 Alphadolone acetate Neurosteriod 22 Alphaprodine Opiod analgesic 23 Alpidem Anxiolytic 24 Alprazolam Anxiolytic 25 Alprenolol Beta blocker 26 Althesin IV anaesthetic 27 Althiazide Diuretic 28 Altrenogest (in males and gelidngs) Oestrus suppression 29 Alverine Antispasmodic 30 Amantadine Dopaminergic 31 Ambenonium Cholinesterase inhibition 32 Ambucetamide Antispasmodic 33 Amethocaine Local anaesthetic 34 Amfepramone Stimulant 35 Amfetaminil Stimulant 36 Amidephrine Vasoconstrictor 37 Amiloride Diuretic 1 Prohibited Substances List This is the Equine Prohibited Substances List that was voted in at the FEI General Assembly in November 2009 alongside the new Equine Anti-Doping and Controlled Medication Regulations(EADCMR). -

Solubilization of the Muscarinic Acetylcholine Receptor by I Sodium Cholate: Stabilization of the Receptor by Muscarinic Hgands
\ i Biomedical Research 3 (6) 695-698, 1982 Solubilization of the muscarinic acetylcholine receptor by I sodium cholate: Stabilization of the receptor by muscarinic Hgands TATSUYA HAGA, TOSHIHIDE NUKADA and KAZUKO HAGA Department of Biochemistry, Hamamatsu University School of Medicine, Hamamatsu 431-31, Japan ABSTRACT Forty to fifty percent of the muscarinic receptor in membrane preparations of porcine caudate nucleus was solubilized by sodium cholate in its active form when the mem- branes were pretreated with carbamylcholine or other muscarinic ligands. The binding activity of solubilized receptors was assayed by using [3H]quinuclidinylben- zylate ([3H]QNB) after removal of cholate and carbamylcholine. Without the pre- treatment, sodium cholate abolished most of the binding activity. The solubilized receptor had a high afiinity for [3H]QNB with a dissociation constant of 58 pM and an affinity for muscarinic ligands similar to that of the membrane receptor. The muscarinic receptor has been studied in —80°C until use. In the standard procedure of detail with respect to its binding with muscarinic solubilization, the microsomal preparation ligands by using [3H]-labeled ligands (3, 12) and (about 5 mg protein per ml) was incubated with several subclasses with different affinities for l0 mM carbamylcholine in 50 mM Tris-HCI agonists (2) or selective antagonists (8) have buffer (pH 7.5) at 30°C for 10 min, then a half been reported. It is not known, however, volume of 3 ‘X, sodium cholate in the Tris buffer whether these subclasses represent several types was added and the incubation continued for 5 of receptors which differ as chemical entities, or min at 30°C. -

Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DIX to the HTSUS—Continued
20558 Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DEPARMENT OF THE TREASURY Services, U.S. Customs Service, 1301 TABLE 1.ÐPHARMACEUTICAL APPEN- Constitution Avenue NW, Washington, DIX TO THE HTSUSÐContinued Customs Service D.C. 20229 at (202) 927±1060. CAS No. Pharmaceutical [T.D. 95±33] Dated: April 14, 1995. 52±78±8 ..................... NORETHANDROLONE. A. W. Tennant, 52±86±8 ..................... HALOPERIDOL. Pharmaceutical Tables 1 and 3 of the Director, Office of Laboratories and Scientific 52±88±0 ..................... ATROPINE METHONITRATE. HTSUS 52±90±4 ..................... CYSTEINE. Services. 53±03±2 ..................... PREDNISONE. 53±06±5 ..................... CORTISONE. AGENCY: Customs Service, Department TABLE 1.ÐPHARMACEUTICAL 53±10±1 ..................... HYDROXYDIONE SODIUM SUCCI- of the Treasury. NATE. APPENDIX TO THE HTSUS 53±16±7 ..................... ESTRONE. ACTION: Listing of the products found in 53±18±9 ..................... BIETASERPINE. Table 1 and Table 3 of the CAS No. Pharmaceutical 53±19±0 ..................... MITOTANE. 53±31±6 ..................... MEDIBAZINE. Pharmaceutical Appendix to the N/A ............................. ACTAGARDIN. 53±33±8 ..................... PARAMETHASONE. Harmonized Tariff Schedule of the N/A ............................. ARDACIN. 53±34±9 ..................... FLUPREDNISOLONE. N/A ............................. BICIROMAB. 53±39±4 ..................... OXANDROLONE. United States of America in Chemical N/A ............................. CELUCLORAL. 53±43±0 -

DIFEN FLEX Eliminated During the First 12 Hours Following Administration
4,5-dihydroxy-diclofenac) represent 10-20% of the dose eliminated in the urine. ® The elimination of diclofenac and its metabolites is rapid: 40% of the administered dose is DIFEN FLEX eliminated during the first 12 hours following administration. Diclofenac pharmacokinetics parameters remain constant after repeated administration in SODIUM DICLOFENAC INJECTABLE SOLUTION healthy subjects. Patients with hepatic dysfunction (chronic hepatitis, cirrhosis without portal PRIDINOL MESYLATE descompensation) show no pharmacokinetics changes compared to healthy subjects. Pridinol Rx Only When Pridinol is administered intravenously to dogs, only 2% of the dose is found in plasma Made in Argentina following injection, less than 9% of the dose is recovered in urine in 2 hours as unchanged pridinol and its glucuronoconjugate. No pridinol is found in urine from 2 to 7 hours after Formulation: each ampoule contains: sodium diclofenac 75.000 mg; pridinol mesylate 2.200 administration. mg. Excipients: monobasic sodium phosphate 5.160 mg; benzyl alcohol 250.000 mg; propylene After oral administration of 14C-pridinol to mice, 94% of the radioactivity leaves the digestive tract glycol 1140.000 mg; sodium metabisulfite 9.000 mg; sodium chloride 17.500 mg; sodium in 12 hours. Maximum plasma radioactivity is detected within 1 hour of administration. Between hydroxide 2.160 mg; hydrochloric acid q.s.; water for injection q.s. 3,000 ml 30 and 40% of the dose is found in bile and tissues, mainly liver and kidneys. The radioactivity is eliminated by 80% at 24 hours and by 96% in 4 days, of which 56% is urinary Therapeutic Action: Analgesic - Anti-inflammatory - Myorelaxant excreted. -

Full-Text (PDF)
African Journal of Pharmacy and Pharmacology Vol. 6(33), pp. 2477-2483, 5 September, 2012 Available online at http://www.academicjournals.org/AJPP DOI: 10.5897/AJPP12.654 ISSN 1996-0816 ©2012 Academic Journals Full Length Research Paper The effect of hydrophobicity/hydrophilicity balance of three kinds of adjuvants on pridinol mesylate orally disintegrating tablets and its quality assessment M. Luo, P. X. Tian, Y. H. Luo*, X. Y. Zhao, Q. Chen and L. Y. Luo College of Pharmaceutical Science, Southwest University, Chongqing 400715, China. Accepted 23 August, 2012 This study was aimed at investigating the effects of 3 kinds of adjuvants on the disintegrating ability of pridinol mesylate orally disintegrating tablets (PMODT), and evaluating the quality of PMODT. The hydrophobic balance of 3 adjuvants (polyvinylpolypyrrolidone (PVPP), microcrystalline cellulose (MCC) and Povidone K30 (PVPK30)) were detected using the L9(34) orthogonal test to determine their effects on the disintegrating ability of PMODT. The quality of PMODT was tested according to the requirements of the Chinese Pharmacopoeia (2010). Results indicated that the disintegrability of PMODT decreased with the increasing amount of PVPP from 4 to 8%. However, no significant influence on the disintegrability was found when the amount of MCC was increased from 10 to 15%. On the other hand, the disintegrability decreased with the increasing amount of MCC when the amount was 15 to 20%. Furthermore, the disintegrability was slightly affected when the amount of PVP K30 was 1 to 2%, while the disintegrability decreased when the amount of PVPK30 was at the range of 2 to 3%. The ratio of hydrophobic / hydrophilic adjuvants affected the disintegrability, which showed a “V” shape curve. -
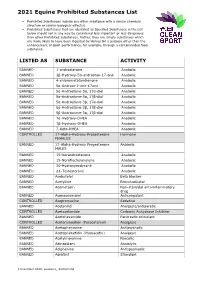
2021 Equine Prohibited Substances List
2021 Equine Prohibited Substances List . Prohibited Substances include any other substance with a similar chemical structure or similar biological effect(s). Prohibited Substances that are identified as Specified Substances in the List below should not in any way be considered less important or less dangerous than other Prohibited Substances. Rather, they are simply substances which are more likely to have been ingested by Horses for a purpose other than the enhancement of sport performance, for example, through a contaminated food substance. LISTED AS SUBSTANCE ACTIVITY BANNED 1-androsterone Anabolic BANNED 3β-Hydroxy-5α-androstan-17-one Anabolic BANNED 4-chlorometatandienone Anabolic BANNED 5α-Androst-2-ene-17one Anabolic BANNED 5α-Androstane-3α, 17α-diol Anabolic BANNED 5α-Androstane-3α, 17β-diol Anabolic BANNED 5α-Androstane-3β, 17α-diol Anabolic BANNED 5α-Androstane-3β, 17β-diol Anabolic BANNED 5β-Androstane-3α, 17β-diol Anabolic BANNED 7α-Hydroxy-DHEA Anabolic BANNED 7β-Hydroxy-DHEA Anabolic BANNED 7-Keto-DHEA Anabolic CONTROLLED 17-Alpha-Hydroxy Progesterone Hormone FEMALES BANNED 17-Alpha-Hydroxy Progesterone Anabolic MALES BANNED 19-Norandrosterone Anabolic BANNED 19-Noretiocholanolone Anabolic BANNED 20-Hydroxyecdysone Anabolic BANNED Δ1-Testosterone Anabolic BANNED Acebutolol Beta blocker BANNED Acefylline Bronchodilator BANNED Acemetacin Non-steroidal anti-inflammatory drug BANNED Acenocoumarol Anticoagulant CONTROLLED Acepromazine Sedative BANNED Acetanilid Analgesic/antipyretic CONTROLLED Acetazolamide Carbonic Anhydrase Inhibitor BANNED Acetohexamide Pancreatic stimulant CONTROLLED Acetominophen (Paracetamol) Analgesic BANNED Acetophenazine Antipsychotic BANNED Acetophenetidin (Phenacetin) Analgesic BANNED Acetylmorphine Narcotic BANNED Adinazolam Anxiolytic BANNED Adiphenine Antispasmodic BANNED Adrafinil Stimulant 1 December 2020, Lausanne, Switzerland 2021 Equine Prohibited Substances List . Prohibited Substances include any other substance with a similar chemical structure or similar biological effect(s). -

United States Patent (19) 11 Patent Number: 6,071,970 Mueller Et Al
USOO607 1970A United States Patent (19) 11 Patent Number: 6,071,970 Mueller et al. (45) Date of Patent: Jun. 6, 2000 54) COMPOUNDS ACTIVE ATA NOVELSITE WOA 93 ON RECEPTOR-OPERATED CALCUM O4373 3/1993 WIPO. CHANNELS USEFUL FOR TREATMENT OF 9521612 8/1995 WIPO. NEUROLOGICAL DISORDERS AND 9605818 2/1996 WIPO. DISEASES 964OO97 12/1996 WIPO. OTHER PUBLICATIONS 75 Inventors: Alan L. Mueller, Salt Lake City; Manuel F. Balandrin, Sandy; Marcusson, Jan O., et al., “Inhibition of Hparoxetine Bradford C. VanWagenen, Salt Lake binding by various Serotonin uptake inhibitors: Structure City; Eric G. DelMar, Salt Lake City; activity-relationships”, European Journal of Pharmacol Scott T. Moe, Salt Lake City; Linda D. ogy, 215, 1992, 191-198. Artman, Salt Lake City; Robert M. Chemical Abstracts, vol. 69, 1968, p. 3322. Barmore, Salt Lake City, all of Utah Chemical Abstracts, vol. 67, 1967, p. 3059. Chemical Abstracts, vol. 66, 1967, p. 4375. 73 Assignee: NPS Pharmaceuticals, Inc., Salt Lake Chemical Abstracts, vol. 5, p. 423, 1959. City, Utah Williams, Ifenprodil Discriminates Subtypes of the N-meth yl-D-asparate Receptor: Selectivity and Mechanisms at 21 Appl. No.: 08/485,038 Recombinant Heteromeric Receptors, Mol. Pharmacol., 44: 851, 1993). 22 Filed: Jun. 7, 1995 Wiley and Balster, Preclinical Evaluation of N-Methyl D-aspartate Antagonists for Antianxiety Effects: a Review Related U.S. Application Data In: Multiple Sigma and PCP Receptor Ligands; Mechanisms 63 Continuation-in-part of application No. PCT/US94/12293, for Neuromodulation and Neuroprotection NPP Books, Ann Oct. 26, 1994, which is a continuation-in-part of application Arbor, Michigan pp. -

Identification of New Functional Inhibitors of Acid Sphingomyelinase
J. Med. Chem. 2008, 51, 219–237 219 Identification of New Functional Inhibitors of Acid Sphingomyelinase Using a Structure-Property-Activity Relation Model Johannes Kornhuber,*,† Philipp Tripal,† Martin Reichel,† Lothar Terfloth,‡ Stefan Bleich,† Jens Wiltfang,† and Erich Gulbins£ Department of Psychiatry and Psychotherapy, UniVersity of Erlangen, Germany, Molecular Networks, Erlangen, Germany, and Department of Molecular Biology, UniVersity of DuisburgsEssen, Germany ReceiVed May 4, 2007 Some organic weak bases induce a detachment from inner lysosomal membranes and subsequent inactivation of acid sphingomyelinase (ASM) and thus work as functional ASM inhibitors. The aim of the present investigation was to develop a structure-property-activity relation (SPAR) model in order to specify the structural and physicochemical characteristics of probes capable of functionally inhibiting ASM. High pKa and high log P values are necessary but not sufficient preconditions for functional inhibition of ASM. The experimental data supported the requirement of an additional factor, which is necessary for functional inhibition of ASM. This factor k is related to the steric hindrance of the most basic nitrogen atom and presumably modulates the free presentation of a protonated nitrogen atom at the inner lysosomal surface. During the course of the study, we characterized 26 new functional ASM inhibitors, including doxepine 63, fluoxetine 104, maprotiline 109, nortriptyline 114, paroxetine 118, sertraline 124, suloctidil 125, and terfenadine 127. Introduction inactivation. Weak bases such as desipramine (17) result in a detachment of the ASM from the inner membrane21 and Eukaryotic cells have the ability to internally segregate subsequent inactivation, possibly by proteolytic degradation.22 aqueous compartments differing in pH using lipid bilayer Weak bases, therefore, do not directly inhibit ASM but result membranes and ATP-driven proton pumps.1–5 The lysosomal in a functional inhibition of ASM.