Plastisphere” Communities†
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(WHO) Report on Microplastics in Drinking Water
Microplastics in drinking-water Microplastics in drinking-water ISBN 978-92-4-151619-8 © World Health Organization 2019 Some rights reserved. This work is available under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons.org/licenses/by-nc-sa/3.0/igo). Under the terms of this licence, you may copy, redistribute and adapt the work for non-commercial purposes, provided the work is appropriately cited, as indicated below. In any use of this work, there should be no suggestion that WHO endorses any specific organization, products or services. The use of the WHO logo is not permitted. If you adapt the work, then you must license your work under the same or equivalent Creative Commons licence. If you create a translation of this work, you should add the following disclaimer along with the suggested citation: “This translation was not created by the World Health Organization (WHO). WHO is not responsible for the content or accuracy of this translation. The original English edition shall be the binding and authentic edition”. Any mediation relating to disputes arising under the licence shall be conducted in accordance with the mediation rules of the World Intellectual Property Organization. Suggested citation. Microplastics in drinking-water. Geneva: World Health Organization; 2019. Licence: CC BY-NC-SA 3.0 IGO. Cataloguing-in-Publication (CIP) data. CIP data are available at http://apps.who.int/iris. Sales, rights and licensing. To purchase WHO publications, see http://apps.who.int/bookorders. To submit requests for commercial use and queries on rights and licensing, see http://www.who.int/about/licensing. -

A Pilot Study to Determine the Potential Impacts of Plastics on Aotearoa-New Zealand's Marine Environment
A pilot study to determine the potential impacts of plastics on Aotearoa-New Zealand's marine environment Olga Pantos∗1, Francois Audrezet2, Fraser Doake1, Lloyd Donaldson3, Pierre Dupont1, Sally Gaw4, Joanne Kingsbury1, Louise Weaver1, Gavin Lear5, Grant Northcott6, Xavier Pochon2,5, Dawn Smith3, Beatrix Theobald3, Jessica Wallbank5, Anastasija Zaiko2,5, and Stefan Maday5 1The Institute of Environmental Science and Research { Christchurch, New Zealand 2Cawthron Institute { Nelson, New Zealand 3Scion { Rotorua, New Zealand 4The University of Canterbury { Christchurch, New Zealand 5University of Auckland [Auckland] { New Zealand 6Northcott Research Consultants Ltd { Hamilton, New Zealand Abstract Once in the ocean, plastics are rapidly colonised by complex communities. Due to the buoyant and resilient nature, ocean plastics pose a significant risk to ecosystems and fishery- based economies through their role in the translocation of invasive species and pathogens or changes in ecosystem function. Factors affecting the development and composition of these communities are still poorly understood, and there is currently no information on the biofilms that form on marine plastics in the southern hemisphere or their potential risks to the environment. This study aims to address this knowledge gap. To do this, two chemically and structurally distinct polymers, which are also common in marine plastic litter, nylon 6 and polyethylene, were deployed for 3-months in the Port of Lyttelton, Christchurch, New Zealand. Biofilm present after 2 weeks was dominated by diatoms and cyanobacteria. Metagenomic analysis showed that the plastisphere was distinct from the communities associated with glass control surfaces and the surrounding water. Polymer-specificity of the bacterial com- munities seen at 2-weeks was absent in subsequent time points, whereas fungal communities did not change over time. -

EASAC Report on Packaging Plastics in the Circular Economy
Packaging plastics in the circular economy Packaging plastics in the circular ea sac Packaging plastics in the circular economy March 2020 March EASAC policy report 39 March 2020 ISBN: 978-3-8047-4129-4 EASAC This report can be found at www.easac.eu Science Advice for the Benefit of Europe EASAC EASAC – the European Academies' Science Advisory Council – is formed by the national science academies of the EU Member States to enable them to collaborate with each other in giving advice to European policy-makers. It thus provides a means for the collective voice of European science to be heard. EASAC was founded in 2001 at the Royal Swedish Academy of Sciences. Its mission reflects the view of academies that science is central to many aspects of modern life and that an appreciation of the scientific dimension is a pre-requisite to wise policy-making. This view already underpins the work of many academies at national level. With the growing importance of the European Union as an arena for policy, academies recognise that the scope of their advisory functions needs to extend beyond the national to cover also the European level. Here it is often the case that a trans-European grouping can be more effective than a body from a single country. The academies of Europe have therefore formed EASAC so that they can speak with a common voice with the goal of building science into policy at EU level. Through EASAC, the academies work together to provide independent, expert, evidence-based advice about the scientific aspects of public policy to those who make or influence policy within the European institutions. -

Hints at the Applicability of Microalgae and Cyanobacteria for the Biodegradation of Plastics
sustainability Review Hints at the Applicability of Microalgae and Cyanobacteria for the Biodegradation of Plastics Giovanni Davide Barone 1,* , Damir Ferizovi´c 2 , Antonino Biundo 3,4 and Peter Lindblad 5 1 Institute of Molecular Biotechnology, Graz University of Technology, 8010 Graz, Austria 2 Institute of Analysis and Number Theory, Graz University of Technology, 8010 Graz, Austria; [email protected] 3 Department of Bioscience, Biotechnology and Biopharmaceutics, University of Bari, 70125 Bari, Italy; [email protected] 4 Interuniversity Consortium for Biotechnology (CIB), 70125 Bari, Italy 5 Department of Chemistry—Ångström Laboratory, Uppsala University, SE-751 20 Uppsala, Sweden; [email protected] * Correspondence: [email protected] Received: 30 October 2020; Accepted: 10 December 2020; Published: 14 December 2020 Abstract: Massive plastic accumulation has been taking place across diverse landscapes since the 1950s, when large-scale plastic production started. Nowadays, societies struggle with continuously increasing concerns about the subsequent pollution and environmental stresses that have accompanied this plastic revolution. Degradation of used plastics is highly time-consuming and causes volumetric aggregation, mainly due to their high strength and bulky structure. The size of these agglomerations in marine and freshwater basins increases daily. Exposure to weather conditions and environmental microflora (e.g., bacteria and microalgae) can slowly corrode the plastic structure. As has been well documented in recent years, plastic fragments are widespread in marine basins and partially in main global rivers. These are potential sources of negative effects on global food chains. Cyanobacteria (e.g., Synechocystis sp. PCC 6803, and Synechococcus elongatus PCC 7942), which are photosynthetic microorganisms and were previously identified as blue-green algae, are currently under close attention for their abilities to capture solar energy and the greenhouse gas carbon dioxide for the production of high-value products. -

UNIVERSITY of CALIFORNIA, SAN DIEGO Abundance and Ecological
UNIVERSITY OF CALIFORNIA, SAN DIEGO Abundance and ecological implications of microplastic debris in the North Pacific Subtropical Gyre A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Oceanography by Miriam Chanita Goldstein Committee in charge: Professor Mark D. Ohman, Chair Professor Lihini I. Aluwihare Professor Brian Goldfarb Professor Michael R. Landry Professor James J. Leichter 2012 Copyright Miriam Chanita Goldstein, 2012 All rights reserved. SIGNATURE PAGE The Dissertation of Miriam Chanita Goldstein is approved, and it is acceptable in quality and form for publication on microfilm and electronically: PAGE _____________________________________________________________________ _____________________________________________________________________ _____________________________________________________________________ _____________________________________________________________________ _____________________________________________________________________ Chair University of California, San Diego 2012 iii DEDICATION For my mother, who took me to the tidepools and didn’t mind my pet earthworms. iv TABLE OF CONTENTS SIGNATURE PAGE ................................................................................................... iii DEDICATION ............................................................................................................. iv TABLE OF CONTENTS ............................................................................................. v LIST OF FIGURES -

The Biogeography of the Plastisphere: Implications for Policy
RESEARCH COMMUNICATIONS RESEARCH COMMUNICATIONS The biogeography of the Plastisphere: 541 implications for policy Linda A Amaral-Zettler1,2*, Erik R Zettler3, Beth Slikas1, Gregory D Boyd3, Donald W Melvin3, Clare E Morrall4, Giora Proskurowski5, and Tracy J Mincer6† Microplastics (particles less than 5 mm) numerically dominate marine debris and occur from coastal waters to mid-ocean gyres, where surface circulation concentrates them. Given the prevalence of plastic marine debris (PMD) and the rise in plastic production, the impacts of plastic on marine ecosystems will likely increase. Microscopic life (the “Plastisphere”) thrives on these tiny floating “islands” of debris and can be transported long distances. Using next-generation DNA sequencing, we characterized bacterial communities from water and plastic samples from the North Pacific and North Atlantic subtropical gyres to determine whether the composition of different Plastisphere communities reflects their biogeographic origins. We found that these communities differed between ocean basins – and to a lesser extent between polymer types – and displayed lat- itudinal gradients in species richness. Our research reveals some of the impacts of microplastics on marine bio- diversity, demonstrates that the effects and fate of PMD may vary considerably in different parts of the global ocean, and suggests that PMD mitigation will require regional management efforts. Front Ecol Environ 2015; 13(10): 541–546, doi:10.1890/150017 ublic awareness of plastic marine debris (PMD) and its Oberbeckmann et al. 2014). As annual plastic produc- Penvironmental impacts has intensified due in part to tion continues to increase (299 million metric tons in greater numbers of publications (both scholarly and popu- 2015 [PlasticsEurope 2015]), citizens, government agen- lar) and large-scale policy recommendations on the sub- cies, and the plastics industry are attempting to manage ject. -

Plastics and the Microbiome: Impacts and Solutions G
Lear et al. Environmental Microbiome (2021) 16:2 Environmental Microbiome https://doi.org/10.1186/s40793-020-00371-w REVIEW Open Access Plastics and the microbiome: impacts and solutions G. Lear1* , J. M. Kingsbury2,S.Franchini1,V.Gambarini1,S.D.M.Maday1,J.A.Wallbank1,L.Weaver2 and O. Pantos2 Abstract Global plastic production has increased exponentially since manufacturing commenced in the 1950’s, including polymer types infused with diverse additives and fillers. While the negative impacts of plastics are widely reported, particularly on marine vertebrates, impacts on microbial life remain poorly understood. Plastics impact microbiomes directly, exerting toxic effects, providing supplemental carbon sources and acting as rafts for microbial colonisation and dispersal. Indirect consequences include increased environmental shading, altered compositions of host communities and disruption of host organism or community health, hormone balances and immune responses. The isolation and application of plastic-degrading microbes are of substantial interest yet little evidence supports the microbial biodegradation of most high molecular weight synthetic polymers. Over 400 microbial species have been presumptively identified as capable of plastic degradation, but evidence for the degradation of highly prevalent polymers including polypropylene, nylon, polystyrene and polyvinyl chloride must be treated with caution; most studies fail to differentiate losses caused by the leaching or degradation of polymer monomers, additives or fillers. Even where polymer degradation is demonstrated, such as for polyethylene terephthalate, the ability of microorganisms to degrade more highly crystalline forms of the polymer used in commercial plastics appears limited. Microbiomes frequently work in conjunction with abiotic factors such as heat and light to impact the structural integrity of polymers and accessibility to enzymatic attack. -

Saving the Mediterranean from Plastic Pollution
REPORT 2018 OUT OF THE PLASTIC TRAP SAVING THE MEDITERRANEAN FROM PLASTIC POLLUTION Front cover A loggerhead sea turtle swims entangled in abandoned CONTENTS fishing gear, off the coast of Tenerife, Canary Islands. World Press Photo 2017. © www.francisperez.es EXECUTIVE SUMMARY 3 1. PLASTICS IN EUROPE 5 Published in June 2018 2. DISTRESS SIGNALS FROM THE SEA 6 by WWF – World Wide Fund For Nature (Formerly World Wildlife Fund). A GLOBAL EMERGENCY 6 Any reproduction in full or in part THE MEDITERRANEAN “PLASTIC TRAP” 10 must mention the title, the lead author, Plastics used and recycled in Mediterranean countries 12 and credit the above-mentioned publisher as the copyright owner. © Text 2018 WWF. All rights reserved 14 Citation of this report: Alessi. et al. 2018. 3. RISKS FOR WILDLIFE “Out of the plastic trap: saving DEADLY TRAPS 14 the Mediterranean from plastic pollution”. JUNK FOOD 15 WWF Mediterranean Marine Initiative, Rome, Italy. 28 pp. Why do animals mistake plastic for food? 17 Lead author: Eva Alessi (WWF) Microplastics alert in the Pelagos sanctuary 18 Co-author: Giuseppe Di Carlo (WWF) SILENT POISONING 19 Communications: Stefania Campogianni (WWF) THE PLASTI-SPHERE 20 Translation: Eda Başgül Di Carlo Editing: Barney Jeffries Design/Layout/Infographics: Bianco Tangerine Snc RECOMMENDATIONS FOR A PLASTIC-FREE MEDITERRANEAN 21 (Erika Vicaretti, Maria Isabella Reggio) This report is available at: ocean.panda.org and mediterranean.panda.org REFERENCES 25 page 2 | Out of the plastic trap. Saving the Mediterranean from plastic pollution EXECUTIVE SUMMARY The Mediterranean Sea, cradle of civilization and centre of extraordinary environmental heritage, is today one of the seas with the highest EUROPE levels of plastic pollution in the world. -

SOURCES, FATE and EFFECTS of MICROPLASTICS in the MARINE ENVIRONMENT: PART 2 of a GLOBAL ASSESSMENT Science for Sustainable Oceans
93 SOURCES, FATE AND EFFECTS OF MICROPLASTICS IN THE MARINE ENVIRONMENT: PART 2 OF A GLOBAL ASSESSMENT Science for Sustainable Oceans ISSN 1020–4873 REPORTS AND STUDIES AND STUDIES REPORTS AND REPORTS 93 SOURCES, FATE AND EFFECTS OF MICROPLASTICS IN THE MARINE ENVIRONMENT: PART TWO OF A GLOBAL ASSESSMENT A report to inform the Second United Nations Environment Assembly GESAMP Working Group 40 2nd phase REPORTS AND STUDIES REPORTS Published by the INTERNATIONAL MARITIME ORGANIZATION 4 Albert Embankment, London SE1 7SR www.imo.org Printed by Micropress Printers Ltd. ISSN: 1020-4873 Cover photo: Peter Kershaw Notes: GESAMP is an advisory body consisting of specialized experts nominated by the Sponsoring Agencies (IMO, FAO, UNESCO-IOC, UNIDO, WMO, IAEA, UN, UNEP, UNDP). Its principal task is to provide scientific advice concerning the prevention, reduction and control of the degradation of the marine environment to the Sponsoring Agencies. The report contains views expressed or endorsed by members of GESAMP who act in their individual capacities; their views may not necessarily correspond with those of the Sponsoring Agencies. Permission may be granted by any of the Sponsoring Agencies for the report to be wholly or partially reproduced in publication by any individual who is not a staff member of a Sponsoring Agency of GESAMP, provided that the source of the extract and the condition mentioned above are indicated. Information about GESAMP and its reports and studies can be found at: http://gesamp.org ISSN 1020-4873 (GESAMP Reports & Studies Series) Copyright © IMO, FAO, UNESCO-IOC, UNIDO, WMO, IAEA, UN, UNEP, UNDP 2015 For bibliographic purposes this document should be cited as: GESAMP (2016). -

Biodegradable Plastics and Marine Litter
www.unep.org United Nations Environment Programme P.O. Box 30552 Nairobi, 00100 Kenya Tel: (254 20) 7621234 Fax: (254 20) 7623927 BIODEGRADABLE E-mail: [email protected] web: www.unep.org PLASTICS & MARINE LITTER MISCONCEPTIONS, CONCERNS AND IMPACTS ON MARINE ENVIRONMENTS 1.002 millimeters 0.0004 millimeters 1.0542 millimeters 1.24 millimeters 0.002 millimeters Copyright © United Nations Environment Programme (UNEP), 2015 This publication may be reproduced in whole or part and in any form for educational or non-profit purposes whatsoever without special permission from the copyright holder, provided that acknowledgement of the source is made. This publication is a contribution to the Global Partnership on Marine Litter (GPML). UNEP acknowledges the financial contribution of the Norwegian government toward the GPML and this publication. Thank you to the editorial reviewers Heidi( Savelli (DEPI, UNEP), Vincent Sweeney (DEPI UNEP), Mette L. Wilkie (DEPI UNEP), Kaisa Uusimaa (DEPI, UNEP), Mick Wilson (DEWA, UNEP) Elisa Tonda (DTIE, UNEP) Ainhoa Carpintero (DTIE/IETC, UNEP) Maria Westerbos, Jeroen Dawos, (Plastic Soup Foundation) Christian Bonten (Universität Stuttgart) Anthony Andrady (North Carolina State University, USA) Author: Dr. Peter John Kershaw Designer: Agnes Rube Cover photo: © Ben Applegarth / Broken Wave Nebula (Creative Commons) © Forest and Kim Starr / Habitat with plastic debris, Hawaii (Creative Commons) ISBN: 978-92-807-3494-2 Job Number: DEP/1908/NA Division of Environmental Policy Implementation Citation: UNEP (2015) Biodegradable Plastics and Marine Litter. Misconceptions, concerns and impacts on marine environments. United Nations Environment Programme (UNEP), Nairobi. Disclaimer The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the United Nations Environment Programme concerning the legal status of any country, territory, city or area or of its authorities, or concerning delimitation of its frontiers or boundaries. -

The Way of Macroplastic Through the Environment
environments Review The Way of Macroplastic through the Environment Simone Lechthaler 1,2,* , Kryss Waldschläger 1 , Georg Stauch 2 and Holger Schüttrumpf 1 1 Institute of Hydraulic Engineering and Water Resources Management, RWTH Aachen University, 52062 Aachen, Germany; [email protected] (K.W.); [email protected] (H.S.) 2 Department of Geography, Physical Geography and Geoecology, RWTH Aachen University, 52062 Aachen, Germany; [email protected] * Correspondence: [email protected] Received: 28 August 2020; Accepted: 22 September 2020; Published: 24 September 2020 Abstract: With the focus on microplastic in current research, macroplastic is often not further considered. Thus, this review paper is the first to analyse the entry paths, accumulation zones, and sinks of macroplastic in the aquatic, terrestrial, and atmospheric environment by presenting transport paths and concentrations in the environment as well as related risks. This is done by applying the Source–Pathway–Receptor model on macroplastic in the environment. Based on this model, the life cycle of macroplastic is structurally described, and knowledge gaps are identified. Hence, current research aspects on macroplastic as well as a sound delimitation between macro- and microplastic that can be applied to future research are indicated. The results can be used as basic information for further research and show a qualitative assessment of the impact of macroplastic that ends up in the environment and accumulates there. Furthermore, the applied model allows for the first time a quantitative and structured approach to macroplastic in the environment. Keywords: macroplastic; life cycle; sources; sinks; transport paths; knowledge gaps 1. -
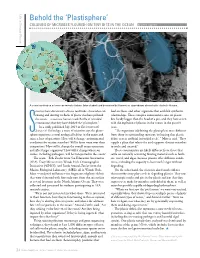
Plastisphere’ COLONIES of MICROBES FLOURISH on TINY BITS in the OCEAN by Lonny Lippsett
Life in Strange Places Behold the ‘Plastisphere’ COLONIES OF MICROBES FLOURISH ON TINY BITS IN THE OCEAN by Lonny Lippsett T o m K l e i n d in s t /W H O I Zettler/SEA, Mincer/WHOI, Amaral-Zettler/MBL A scanning electron microscope reveals diatoms (blue shaded) and bacteria with filamentous appendages aboard rafts of plastic flotsam. cientists have discovered a diverse multitude of microbes col- feed on these, and other organisms that establish symbiotic onizing and thriving on flecks of plastic that have polluted relationships. These complex communities exist on plastic the oceans—a vast new human-made flotilla of microbial bits hardly bigger than the head of a pin, and they have arisen communities that they have dubbed the “plastisphere.” with the explosion of plastics in the oceans in the past 60 In a study published July 2013 in Environmental years. Science & Technology, a team of scientists says the plasti- “The organisms inhabiting the plastisphere were different Ssphere represents a novel ecological habitat in the ocean and from those in surrounding seawater, indicating that plastic raises a host of questions: How will it change environmental debris acts as artificial ‘microbial reefs,’ ” Mincer said. “They conditions for marine microbes? Will it favor some over their supply a place that selects for and supports distinct microbes competitors? How will it change the overall ocean ecosystem to settle and succeed.” and affect larger organisms? How will it change where mi- These communities are likely different from those that crobes, including pathogens, will be transported in the ocean? settle on naturally occurring floating material such as feath- The team—Erik Zettler from Sea Education Association ers, wood, and algae, because plastics offer different condi- (SEA), Tracy Mincer from Woods Hole Oceanographic tions, including the capacity to last much longer without Institution (WHOI), and Linda Amaral-Zettler from the degrading.