PDF (Supplementary Figures)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification and Developmental Expression of the Full Complement Of
Goldstone et al. BMC Genomics 2010, 11:643 http://www.biomedcentral.com/1471-2164/11/643 RESEARCH ARTICLE Open Access Identification and developmental expression of the full complement of Cytochrome P450 genes in Zebrafish Jared V Goldstone1, Andrew G McArthur2, Akira Kubota1, Juliano Zanette1,3, Thiago Parente1,4, Maria E Jönsson1,5, David R Nelson6, John J Stegeman1* Abstract Background: Increasing use of zebrafish in drug discovery and mechanistic toxicology demands knowledge of cytochrome P450 (CYP) gene regulation and function. CYP enzymes catalyze oxidative transformation leading to activation or inactivation of many endogenous and exogenous chemicals, with consequences for normal physiology and disease processes. Many CYPs potentially have roles in developmental specification, and many chemicals that cause developmental abnormalities are substrates for CYPs. Here we identify and annotate the full suite of CYP genes in zebrafish, compare these to the human CYP gene complement, and determine the expression of CYP genes during normal development. Results: Zebrafish have a total of 94 CYP genes, distributed among 18 gene families found also in mammals. There are 32 genes in CYP families 5 to 51, most of which are direct orthologs of human CYPs that are involved in endogenous functions including synthesis or inactivation of regulatory molecules. The high degree of sequence similarity suggests conservation of enzyme activities for these CYPs, confirmed in reports for some steroidogenic enzymes (e.g. CYP19, aromatase; CYP11A, P450scc; CYP17, steroid 17a-hydroxylase), and the CYP26 retinoic acid hydroxylases. Complexity is much greater in gene families 1, 2, and 3, which include CYPs prominent in metabolism of drugs and pollutants, as well as of endogenous substrates. -

Synonymous Single Nucleotide Polymorphisms in Human Cytochrome
DMD Fast Forward. Published on February 9, 2009 as doi:10.1124/dmd.108.026047 DMD #26047 TITLE PAGE: A BIOINFORMATICS APPROACH FOR THE PHENOTYPE PREDICTION OF NON- SYNONYMOUS SINGLE NUCLEOTIDE POLYMORPHISMS IN HUMAN CYTOCHROME P450S LIN-LIN WANG, YONG LI, SHU-FENG ZHOU Department of Nutrition and Food Hygiene, School of Public Health, Peking University, Beijing 100191, P. R. China (LL Wang & Y Li) Discipline of Chinese Medicine, School of Health Sciences, RMIT University, Bundoora, Victoria 3083, Australia (LL Wang & SF Zhou). 1 Copyright 2009 by the American Society for Pharmacology and Experimental Therapeutics. DMD #26047 RUNNING TITLE PAGE: a) Running title: Prediction of phenotype of human CYPs. b) Author for correspondence: A/Prof. Shu-Feng Zhou, MD, PhD Discipline of Chinese Medicine, School of Health Sciences, RMIT University, WHO Collaborating Center for Traditional Medicine, Bundoora, Victoria 3083, Australia. Tel: + 61 3 9925 7794; fax: +61 3 9925 7178. Email: [email protected] c) Number of text pages: 21 Number of tables: 10 Number of figures: 2 Number of references: 40 Number of words in Abstract: 249 Number of words in Introduction: 749 Number of words in Discussion: 1459 d) Non-standard abbreviations: CYP, cytochrome P450; nsSNP, non-synonymous single nucleotide polymorphism. 2 DMD #26047 ABSTRACT Non-synonymous single nucleotide polymorphisms (nsSNPs) in coding regions that can lead to amino acid changes may cause alteration of protein function and account for susceptivity to disease. Identification of deleterious nsSNPs from tolerant nsSNPs is important for characterizing the genetic basis of human disease, assessing individual susceptibility to disease, understanding the pathogenesis of disease, identifying molecular targets for drug treatment and conducting individualized pharmacotherapy. -

CYP2F1 (NM 000774) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC223097 CYP2F1 (NM_000774) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: CYP2F1 (NM_000774) Human Tagged ORF Clone Tag: Myc-DDK Symbol: CYP2F1 Synonyms: C2F1; CYP2F; CYPIIF1 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 4 CYP2F1 (NM_000774) Human Tagged ORF Clone – RC223097 ORF Nucleotide >RC223097 representing NM_000774 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGGACAGCATAAGCACAGCCATCTTACTCCTGCTCCTGGCTCTCGTCTGTCTGCTCCTGACCCTAAGCT CAAGAGATAAGGGAAAGCTGCCTCCGGGACCCAGACCCCTCTCAATCCTGGGAAACCTGCTGCTGCTTTG CTCCCAAGACATGCTGACTTCTCTCACTAAGCTGAGCAAGGAGTATGGCTCCATGTACACAGTGCACCTG GGACCCAGGCGGGTGGTGGTCCTCAGCGGGTACCAAGCTGTGAAGGAGGCCCTGGTGGACCAGGGAGAGG AGTTTAGTGGCCGCGGTGACTACCCTGCCTTTTTCAACTTTACCAAGGGCAATGGCATCGCCTTCTCCAG TGGGGATCGATGGAAGGTCCTGAGACAGTTCTCTATCCAGATTCTACGGAATTTCGGGATGGGGAAGAGA AGCATTGAGGAGCGAATCCTAGAGGAGGGCAGCTTCCTGCTGGCGGAGCTGCGGAAAACTGAAGGCGAGC CCTTTGACCCCACGTTTGTGCTGAGTCGCTCAGTGTCCAACATTATCTGTTCCGTGCTCTTCGGCAGCCG CTTCGACTATGATGATGAGCGTCTGCTCACCATTATCCGCCTTATCAATGACAACTTCCAAATCATGAGC -

Metabolic Activation and Toxicological Evaluation of Polychlorinated Biphenyls in Drosophila Melanogaster T
www.nature.com/scientificreports OPEN Metabolic activation and toxicological evaluation of polychlorinated biphenyls in Drosophila melanogaster T. Idda1,7, C. Bonas1,7, J. Hofmann1, J. Bertram1, N. Quinete1,2, T. Schettgen1, K. Fietkau3, A. Esser1, M. B. Stope4, M. M. Leijs3, J. M. Baron3, T. Kraus1, A. Voigt5,6 & P. Ziegler1* Degradation of polychlorinated biphenyls (PCBs) is initiated by cytochrome P450 (CYP) enzymes and includes PCB oxidation to OH-metabolites, which often display a higher toxicity than their parental compounds. In search of an animal model refecting PCB metabolism and toxicity, we tested Drosophila melanogaster, a well-known model system for genetics and human disease. Feeding Drosophila with lower chlorinated (LC) PCB congeners 28, 52 or 101 resulted in the detection of a human-like pattern of respective OH-metabolites in fy lysates. Feeding fies high PCB 28 concentrations caused lethality. Thus we silenced selected CYPs via RNA interference and analyzed the efect on PCB 28-derived metabolite formation by assaying 3-OH-2′,4,4′-trichlorobiphenyl (3-OHCB 28) and 3′-OH-4′,4,6′-trichlorobiphenyl (3′-OHCB 28) in fy lysates. We identifed several drosophila CYPs (dCYPs) whose knockdown reduced PCB 28-derived OH-metabolites and suppressed PCB 28 induced lethality including dCYP1A2. Following in vitro analysis using a liver-like CYP-cocktail, containing human orthologues of dCYP1A2, we confrm human CYP1A2 as a PCB 28 metabolizing enzyme. PCB 28-induced mortality in fies was accompanied by locomotor impairment, a common phenotype of neurodegenerative disorders. Along this line, we show PCB 28-initiated caspase activation in diferentiated fy neurons. -

Strong Correlation Between Air-Liquid Interface Cultures and in Vivo Transcriptomics of Nasal Brush Biopsy
Am J Physiol Lung Cell Mol Physiol 318: L1056–L1062, 2020. First published April 1, 2020; doi:10.1152/ajplung.00050.2020. RAPID REPORT Translational Physiology Strong correlation between air-liquid interface cultures and in vivo transcriptomics of nasal brush biopsy X Baishakhi Ghosh,1 Bongsoo Park,1 Debarshi Bhowmik,2 Kristine Nishida,3 Molly Lauver,3 Nirupama Putcha,3 Peisong Gao,4 Murugappan Ramanathan, Jr.,5 Nadia Hansel,3 Shyam Biswal,1 and Venkataramana K. Sidhaye1,3 1Department of Environmental Health and Engineering, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland; 2Department of Biology, Johns Hopkins University, Baltimore, Maryland; 3Department of Pulmonary and Critical Care Medicine, Johns Hopkins School of Medicine, Baltimore, Maryland; 4Division of Allergy and Clinical Immunology, Johns Hopkins School of Medicine, Baltimore, Maryland; and 5Department of Otolaryngology-Head and Neck Surgery, Johns Hopkins School of Medicine, Baltimore, Maryland Submitted 18 February 2020; accepted in final form 24 March 2020 Ghosh B, Park B, Bhowmik D, Nishida K, Lauver M, Putcha N, RNA sequencing to determine the correlation between ALI- Gao P, Ramanathan M Jr, Hansel N, Biswal S, Sidhaye VK. cultured epithelial cells and epithelial cells obtained from nasal Strong correlation between air-liquid interface cultures and in vivo brushing and identify differences that may arise as a result of transcriptomics of nasal brush biopsy. Am J Physiol Lung Cell Mol redifferentiation. Physiol 318: L1056–L1062, 2020. First published April 1, 2020; doi:10.1152/ajplung.00050.2020.—Air-liquid interface (ALI) cultures METHODS are ex vivo models that are used extensively to study the epithelium of patients with chronic respiratory diseases. -

Expression of CYP2S1 in Human Hepatic Stellate Cells
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector FEBS Letters 581 (2007) 781–786 Expression of CYP2S1 in human hepatic stellate cells Carylyn J. Mareka, Steven J. Tuckera, Matthew Korutha, Karen Wallacea,b, Matthew C. Wrighta,b,* a School of Medical Sciences, Institute of Medical Science, University of Aberdeen, Aberdeen, UK b Liver Faculty Research Group, School of Clinical and Laboratory Sciences, University of Newcastle, Newcastle, UK Received 22 November 2006; revised 16 January 2007; accepted 23 January 2007 Available online 2 February 2007 Edited by Laszlo Nagy the expression and accumulation of scarring extracellular Abstract Activated stellate cells are myofibroblast-like cells associated with the generation of fibrotic scaring in chronically fibrotic matrix protein [2]. It is currently thought an inhibition damaged liver. Gene chip analysis was performed on cultured fi- of fibrosis in liver diseases may be an effective approach to brotic stellate cells. Of the 51 human CYP genes known, 13 treating patients for which the cause is refractive to current CYP and 5 CYP reduction-related genes were detected with 4 treatments (e.g. in approx. 30% of hepatitis C infected CYPs (CYP1A1, CYP2E1, CY2S1 and CYP4F3) consistently patients) [2,3]. At present, there is no approved treatment for present in stellate cells isolated from three individuals. Quantita- fibrosis. tive RT-PCR indicated that CYP2S1 was a major expressed Inadvertent toxicity of drugs is often associated with a ‘‘met- CYP mRNA transcript. The presence of a CYP2A-related pro- abolic activation’’ by CYPs [1]. -
CYTOCHROME P450: Structure-Function
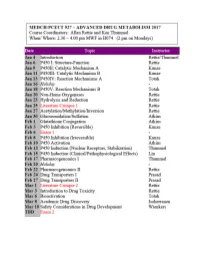
MEDCH 527 AER Jan. 4-6, 2017 CYTOCHROME P450: Structure-Function 1. General P450 Characteristics anD Taxonomy 2. Human P450s – Substrate anD Inhibitor Selectivities 3. Structure-Function Aspects of LiganD BinDing, P450 ReDuction anD Oxygen Activation References P450 Homepage -http:// http://drnelson.uthsc.edu/CytochromeP450.html Nelson DR et al., Pharmacogenetics. 1996 6(1):1-42: Nelson DR et al., Pharmacogenetics. 2004 14(1):1-18. Testa, B. The Biochemistry of Drug Metabolism: A 6 Part Series in Chem. BioDivers. (2006-2008). Sligar, SG. Glimpsing the critical intermediate in cytochrome P450 oxidations. Science. 2010 Nov 12;330(6006):924-5. Johnson EF, et al. Correlating structure and function of drug metabolizing enzymes:Progress and ongoing challenges. Drug Metab. Dispos. 42:9-22 (2014). Zientek MA and Youdim K, Reaction Phenotyping:Advances in experimental strategies used to characterize the contribution of drug metabolizing enzymes. Drug Metab. Dispos. 43:163-181 (2015). Almazroo et al., Drug Metabolism in the Liver. Clin. Liver Dis. 21:1-20 (2017). Absorption, Distribution, Metabolism and Excretion (ADME) of Orally Administered Drugs To reach their sites of action in the body, orally administered drugs must be absorbed from the small intestine, survive first pass metabolism - typically in the liver - before eventually being excreted, usually as drug metabolites in the bile and kidney. Therefore, clinically useful drugs must be able to cross an array of cell membranes, which are composed of a lipid bilayer. Drugs must exhibit an adequate degree of lipophilicity (logP of ~2-4) in order to able to dissolve into this lipoidal environment. Many drug metabolism processes render lipophilic drugs more water-soluble so as to facilitate excretion via the kidneys and bile. -

Sputum Proteomics and Airway Cell Transcripts of Current and Ex-Smokers with Severe Asthma in U-BIOPRED
Sputum proteomics and airway cell transcripts of current and ex-smokers with severe asthma in U-BIOPRED: an exploratory analysis Kentaro Takahashi 1,2, Stelios Pavlidis3, Francois Ng Kee Kwong 1, Uruj Hoda 1, Christos Rossios1, Kai Sun3, Matthew Loza4, Fred Baribaud4, Pascal Chanez5, Steve J Fowler6, Ildiko Horvath7, Paolo Montuschi8, Florian Singer9, Jacek Musial10, Barbro Dahlen11, Sven-Eric Dahlen11, N. Krug12, Thomas Sandstrom13, Dominic E. Shaw14, Rene Lutter 15, Per Bakke16, Louise J. Fleming1, Peter H. Howarth17, Massimo Caruso18, Ana R Sousa19, Julie Corfield20, Charles Auffray21, Bertrand De Meulder21, Diane Lefaudeux21, Ratko Djukanovic17, Peter J Sterk16, Yike Guo3, Ian M. Adcock1,3, Kian Fan Chung1,3 on behalf of the U-BIOPRED study group# 1National Heart & Lung Institute, Imperial College London, & Biomedical Research Unit, Biomedical Research Unit, Royal Brompton & Harefield NHS Trust, London, United Kingdom; 2Research Centre for Allergy and Clinical Immunology, Asahi General Hospital, Japan; 3Department of Computing & Data Science Institute, Imperial College London, United Kingdom; 4Janssen Research and Development, High Wycombe, Buckinghamshire, United Kingdom; 5 Assistance Publique des Hôpitaux de Marseille - Clinique des bronches, allergies et sommeil, Aix Marseille Université, Marseille, France 6 Centre for Respiratory Medicine and Allergy, Institute of Inflammation and Repair, University of Manchester and University Hospital of South Manchester, Manchester Academic Health Sciences Centre, Manchester, United Kingdom -

Inhibition of Cytochrome P450 Enzymes
7 Inhibition of Cytochrome P450 Enzymes Maria Almira Correia and Paul R. Ortiz de Monteflano 1. Introduction of P450 inhibitors are available in various reviews"^^^. This chapter focuses on the mecha Three steps in the catalytic cycle of nisms of inactivation; thus, most of the chapter is cytochrome P450 (P450, CYP; see Chapters 5 and devoted to the discussion of agents that require 6) are particularly vulnerable to inhibition: (a) the P450 catalysis to fiilfill their inhibitory potential. binding of substrates, (b) the binding of molecular The mechanisms of reversible competitive and oxygen subsequent to the first electron transfer, noncompetitive inhibitors, despite their practical and (c) the catalytic step in which the substrate is importance, are relatively straightforward and are actually oxidized. Only inhibitors that act at one of discussed more briefly. these three steps will be considered in this chapter. Inhibitors that act at other steps in the catalytic cycle, such as agents that interfere with the 2. Reversible Inhibitors electron supply to the hemoprotein by accepting electrons directly from P450 reductase^"^, are not Reversible inhibitors compete with substrates discussed here. for occupancy of the active site and include agents P450 inhibitors can be divided into three that (a) bind to hydrophobic regions of the active mechanistically distinct classes: Agents that site, (b) coordinate to the heme iron atom, or (a) bind reversibly, (b) form quasi-irreversible (c) enter into specific hydrogen bonding or ionic complexes with the heme iron atom, and (c) bind interactions with active-site residues"*"^^. The first irreversibly to the protein or the heme moiety, or mechanism, in which the inhibitor simply competes accelerate the degradation and/or oxidative frag for binding to lipophilic domains of the active site, mentation of the prosthetic heme. -

CYP2F1 Rabbit Pab
Leader in Biomolecular Solutions for Life Science CYP2F1 Rabbit pAb Catalog No.: A7550 Basic Information Background Catalog No. This gene encodes a member of the cytochrome P450 superfamily of enzymes. The A7550 cytochrome P450 proteins are monooxygenases which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. This protein Observed MW localizes to the endoplasmic reticulum and is known to dehydrogenate 3-methylindole, 46-54kDa an endogenous toxin derived from the fermentation of tryptophan, as well as xenobiotic substrates such as naphthalene and ethoxycoumarin. This gene is part of a large cluster Calculated MW of cytochrome P450 genes from the CYP2A, CYP2B and CYP2F subfamilies on 36kDa/55kDa chromosome 19q. Category Primary antibody Applications WB, IHC Cross-Reactivity Human, Mouse, Rat Recommended Dilutions Immunogen Information WB 1:500 - 1:2000 Gene ID Swiss Prot 1572 P24903 IHC 1:50 - 1:200 Immunogen Recombinant fusion protein containing a sequence corresponding to amino acids 100-290 of human CYP2F1 (NP_000765.2). Synonyms CYP2F1;C2F1;CYP2F;CYPIIF1 Contact Product Information www.abclonal.com Source Isotype Purification Rabbit IgG Affinity purification Storage Store at -20℃. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide,50% glycerol,pH7.3. Validation Data Western blot analysis of extracts of various cell lines, using CYP2F1 antibody (A7550) at 1:1000 dilution. Secondary antibody: HRP Goat Anti-Rabbit IgG (H+L) (AS014) at 1:10000 dilution. Lysates/proteins: 25ug per lane. Blocking buffer: 3% nonfat dry milk in TBST. Detection: ECL Basic Kit (RM00020). Exposure time: 90s. Immunohistochemistry of paraffin- embedded mouse lung using CYP2F1 antibody (A7550) at dilution of 1:200 (40x lens). -

Profiling Cytochrome P450 Expression in Ovarian Cancer: Identification of Prognostic Markers Diane Downie,1Morag C.E
Imaging, Diagnosis, Prognosis Profiling Cytochrome P450 Expression in Ovarian Cancer: Identification of Prognostic Markers Diane Downie,1Morag C.E. McFadyen,1Patrick H. Rooney,1, 3 Margaret E. Cruickshank,2 David E. Parkin,2 Iain D. Miller,1Colin Telfer,3 William T. Melvin,3 and Graeme I. Murray1 Abstract Purpose: The cytochromes P450 are a multigene family of enzymes with a central role in the oxidative metabolism of a wide range of xenobiotics, including anticancer drugs and biologically active endogenous compounds. The purpose of this study was to define the cytochrome P450 profile of ovarian cancer and identify novel therapeutic targets and establish the prognostic signif- icance of expression of individualcytochrome P450s in this type of cancer. Experimental Design: Immunohistochemistry for a panelof 23 cytochrome P450s and cyto- chrome P450 reductase was done on an ovarian cancer tissue microarray consisting of 99primary epithelialovarian cancers, 22 peritonealmetastasis, and 13 normalovarian samples.The intensity of immunoreactivity in each sample was established by light microscopy. Results: In primary ovarian cancer, several P450s (CYP1B1, CYP2A/2B, CYP2F1, CYP2R1, CYP2U1,CYP3A5, CYP3A7,CYP3A43, CYP4Z1,CYP26A1,and CYP51)were present at a signif- icantlyhigher levelof intensity compared with normalovary. P450 expression was alsodetected in ovarian cancer metastasis and CYP2S1and P450 reductase both showed significantly in- creased expression in metastasis compared with primary ovarian cancer. The presence of low/ negative CYP2A/2B (log rank = 7.06, P = 0.008) or positive CYP4Z1 (log rank = 6.19, P =0.01) immunoreactivity in primary ovarian cancer were each associated with poor prognosis. Both CYP2A/2B and CYP4Z1were also independent markers of prognosis. -

Biodiversity of P-450 Monooxygenase: Cross-Talk
Cytochrome P450: Oxygen activation and biodiversty 1 Biodiversity of P-450 monooxygenase: Cross-talk between chemistry and biology Heme Fe(II)-CO complex 450 nm, different from those of hemoglobin and other heme proteins 410-420 nm. Cytochrome Pigment of 450 nm Cytochrome P450 CYP3A4…. 2 High Energy: Ultraviolet (UV) Low Energy: Infrared (IR) Soret band 420 nm or g-band Mb Fe(II) ---------- Mb Fe(II) + CO - - - - - - - Visible region Visible bands Q bands a-band, b-band b a 3 H2O/OH- O2 CO Fe(III) Fe(II) Fe(II) Fe(II) Soret band at 420 nm His His His His metHb deoxy Hb Oxy Hb Carbon monoxy Hb metMb deoxy Mb Oxy Mb Carbon monoxy Mb H2O/Substrate O2-Substrate CO Substrate Soret band at 450 nm Fe(III) Fe(II) Fe(II) Fe(II) Cytochrome P450 Cys Cys Cys Cys Active form 4 Monooxygenase Reactions by Cytochromes P450 (CYP) + + RH + O2 + NADPH + H → ROH + H2O + NADP RH: Hydrophobic (lipophilic) compounds, organic compounds, insoluble in water ROH: Less hydrophobic and slightly soluble in water. Drug metabolism in liver ROH + GST → R-GS GST: glutathione S-transferase ROH + UGT → R-UG UGT: glucuronosyltransferaseGlucuronic acid Insoluble compounds are converted into highly hydrophilic (water soluble) compounds. 5 Drug metabolism at liver: Sleeping pill, pain killer (Narcotic), carcinogen etc. Synthesis of steroid hormones (steroidgenesis) at adrenal cortex, brain, kidney, intestine, lung, Animal (Mammalian, Fish, Bird, Insect), Plants, Fungi, Bacteria 6 NSAID: non-steroid anti-inflammatory drug 7 8 9 10 11 Cytochrome P450: Cysteine-S binding to Fe(II) heme is important for activation of O2.