1 the Circulating SARS-Cov-2 Spike Variant N439K Maintains
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Baby Girl Names Registered in 2018
Page 1 of 46 Baby Girl Names Registered in 2018 Frequency Name Frequency Name Frequency Name 8 Aadhya 1 Aayza 1 Adalaide 1 Aadi 1 Abaani 2 Adalee 1 Aaeesha 1 Abagale 1 Adaleia 1 Aafiyah 1 Abaigeal 1 Adaleigh 4 Aahana 1 Abayoo 1 Adalia 1 Aahna 2 Abbey 13 Adaline 1 Aaila 4 Abbie 1 Adallynn 3 Aaima 1 Abbigail 22 Adalyn 3 Aaira 17 Abby 1 Adalynd 1 Aaiza 1 Abbyanna 1 Adalyne 1 Aaliah 1 Abegail 19 Adalynn 1 Aalina 1 Abelaket 1 Adalynne 33 Aaliyah 2 Abella 1 Adan 1 Aaliyah-Jade 2 Abi 1 Adan-Rehman 1 Aalizah 1 Abiageal 1 Adara 1 Aalyiah 1 Abiela 3 Addalyn 1 Aamber 153 Abigail 2 Addalynn 1 Aamilah 1 Abigaille 1 Addalynne 1 Aamina 1 Abigail-Yonas 1 Addeline 1 Aaminah 3 Abigale 2 Addelynn 1 Aanvi 1 Abigayle 3 Addilyn 2 Aanya 1 Abiha 1 Addilynn 1 Aara 1 Abilene 66 Addison 1 Aaradhya 1 Abisha 3 Addisyn 1 Aaral 1 Abisola 1 Addy 1 Aaralyn 1 Abla 9 Addyson 1 Aaralynn 1 Abraj 1 Addyzen-Jerynne 1 Aarao 1 Abree 1 Adea 2 Aaravi 1 Abrianna 1 Adedoyin 1 Aarcy 4 Abrielle 1 Adela 2 Aaria 1 Abrienne 25 Adelaide 2 Aariah 1 Abril 1 Adelaya 1 Aarinya 1 Abrish 5 Adele 1 Aarmi 2 Absalat 1 Adeleine 2 Aarna 1 Abuk 1 Adelena 1 Aarnavi 1 Abyan 2 Adelin 1 Aaro 1 Acacia 5 Adelina 1 Aarohi 1 Acadia 35 Adeline 1 Aarshi 1 Acelee 1 Adéline 2 Aarushi 1 Acelyn 1 Adelita 1 Aarvi 2 Acelynn 1 Adeljine 8 Aarya 1 Aceshana 1 Adelle 2 Aaryahi 1 Achai 21 Adelyn 1 Aashvi 1 Achan 2 Adelyne 1 Aasiyah 1 Achankeng 12 Adelynn 1 Aavani 1 Achel 1 Aderinsola 1 Aaverie 1 Achok 1 Adetoni 4 Aavya 1 Achol 1 Adeyomola 1 Aayana 16 Ada 1 Adhel 2 Aayat 1 Adah 1 Adhvaytha 1 Aayath 1 Adahlia 1 Adilee 1 -

What's in an Irish Name?
What’s in an Irish Name? A Study of the Personal Naming Systems of Irish and Irish English Liam Mac Mathúna (St Patrick’s College, Dublin) 1. Introduction: The Irish Patronymic System Prior to 1600 While the history of Irish personal names displays general similarities with the fortunes of the country’s place-names, it also shows significant differences, as both first and second names are closely bound up with the ego-identity of those to whom they belong.1 This paper examines how the indigenous system of Gaelic personal names was moulded to the requirements of a foreign, English-medium administration, and how the early twentieth-century cultural revival prompted the re-establish- ment of an Irish-language nomenclature. It sets out the native Irish system of surnames, which distinguishes formally between male and female (married/ un- married) and shows how this was assimilated into the very different English sys- tem, where one surname is applied to all. A distinguishing feature of nomen- clature in Ireland today is the phenomenon of dual Irish and English language naming, with most individuals accepting that there are two versions of their na- me. The uneasy relationship between these two versions, on the fault-line of lan- guage contact, as it were, is also examined. Thus, the paper demonstrates that personal names, at once the pivots of individual and group identity, are a rich source of continuing insight into the dynamics of Irish and English language contact in Ireland. Irish personal names have a long history. Many of the earliest records of Irish are preserved on standing stones incised with the strokes and dots of ogam, a 1 See the paper given at the Celtic Englishes II Colloquium on the theme of “Toponyms across Languages: The Role of Toponymy in Ireland’s Language Shifts” (Mac Mathúna 2000). -
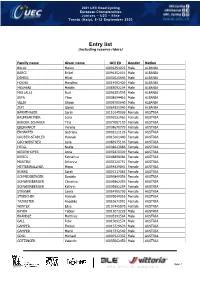
Entry List (Including Reserve Riders)
2021 UEC Road Cycling European Championships Juniors – U23 – Elite Trento (Italy), 8-12 September 2021 Entry list (including reserve riders) Family name Given name UCI ID Gender Nation BALAJ Mateo 10096354023 Male ALBANIA BARCI Brikel 10096352003 Male ALBANIA DEMIRI Mikel 10032025643 Male ALBANIA HOXHA Marolino 10014592420 Male ALBANIA MALHANI Mejdin 10083092204 Male ALBANIA MULLALLI Nuri 10096351595 Male ALBANIA SEFA Ylber 10008694416 Male ALBANIA VELIA Olsian 10009705640 Male ALBANIA ZEFI Gjergj 10064821040 Male ALBANIA BÄRNTHALER Sarah 10110145096 Female AUSTRIA BAUMGARTNER Lena 10096522963 Female AUSTRIA BERGER-SCHAUER Tina 10077087193 Female AUSTRIA EBERHARDT Verena 10008670770 Female AUSTRIA ERHARTER Gabriela 10092222126 Female AUSTRIA GRUBER-STADLER Hannah 10015661440 Female AUSTRIA GSCHWENTNER Leila 10083975106 Female AUSTRIA HEIGL Nadja 10008623886 Female AUSTRIA KIESENHOFER Anna 10092870309 Female AUSTRIA KREIDL Katharina 10048898084 Female AUSTRIA MARTINI Johanna 10035120751 Female AUSTRIA MITTERWALLNER Mona 10054839841 Female AUSTRIA RIJKES Sarah 10007217083 Female AUSTRIA SCHMIDSBERGER Daniela 10099849356 Female AUSTRIA SCHWEINBERGER Christina 10009862355 Female AUSTRIA SCHWEINBERGER Kathrin 10009862254 Female AUSTRIA STIGGER Laura 10054955736 Female AUSTRIA STREICHER Hannah 10035049316 Female AUSTRIA TAZREITER Angelika 10010671091 Female AUSTRIA WINTER Elisa 10107445870 Female AUSTRIA BAYER Tobias 10011072229 Male AUSTRIA BRÄNDLE Matthias 10005391564 Male AUSTRIA GALL Felix 10015092574 Male AUSTRIA GAMPER Florian 10015329620 -

Sainsbury's Féile
First Name Last Name 1 Aaron McCoy 2 Adele Burns 3 Adrian McMullan 4 Aidan Darragh 5 Aidan Doyle 6 Aidan O'Neill 7 Aidan Donnelly 8 Áine Scullion 9 Aisling Mc Guinness 10 Alan Hanna 11 Alan Black 12 Alice Scott 13 Alison O'Rysdale 14 Alison Finnegan 15 Alison McCaughley 16 Amanda Carleton 17 Andrew Johnston 18 Andy Fennell 19 Angela Jackson 20 Ann-Marie McCoy 21 Anna Clugston 22 Anne Caldwell 23 Anne Gribbon 24 Annette Loughran 25 Annette Hinchcliffe 26 Anthony Todd 27 Barney O;Loughlinn 28 Barry McCafferty 29 Barry Corscaden 30 Bernadette Murray 31 Bert McKay 32 Billy McLarnon 33 Breige Flynn 34 Breige Black 35 Brendan Mulgrew 36 Brendan Grew 37 Brian Coney 38 Brian Murray 39 Brigid Moore 40 Bronach Ní Tuama 41 Bronagh Blaney 42 Caitriona Harkin 43 Caoimhe O'Neil 44 Carl McKeating 45 Carol Gibson 46 Carol Walsh 47 Caroline Palmer 48 Catherine White 49 Catherine McLoughlin 50 Catherine Regan 51 Cathy Shields 52 Cathy Gribbon 53 Chris Donnelly 54 Chris Hart 55 Chris Ward 56 Chrissy Dougal 57 Christopher McAvoy 58 Ciara Dolan 59 Ciara Gallagher 60 Ciara Quinn 61 Ciara McGarry 62 Ciara MacKenzie 63 Ciaran Reilly 64 Claire McKeown 65 Claire Palmer 66 Claire McDonagh 67 Clare Holmes 68 Clodagh Delaney 69 Colette McCartney 70 Colette Bailey 71 Colin Cooney 72 Colin Drain 73 Colleen Hillick 74 Colum Benstead 75 Conal Corr 76 Connolly O'Connor 77 Conor McCaffrey 78 Conor Diver 79 Conor Agnew 80 Conor Martin 81 Cuan O'Neill 82 Cutis Millen 83 Damian McCorry 84 Darren Hesketh 85 David McGimpsey 86 David Kearney 87 Deaglan Black 88 Debbie Doherty -

Our Lady's Bulletin
Volume 11 Issue 1 Our Lady’s Bulletin Introduction It is hard to believe that we are half way through the first and longest term of the school year. Already so much has happened as you will see from this Bulletin. I want to take this opportunity to congratulate our Leaving Certificate Class of 2012 on truly fantastic results in August and wish them the very best in their new life paths. A special word of congratulations to Frances Ryan & Fearghal Donaghy who have been awarded the very prestigious Naughton Scholarships. Also our Junior Cert Class really surpassed themselves and they too deserve all our congratulations. A highlight of the new year so far has been the number of new teachers who started with us in September. I want to warmly welcome them all to Our Lady’s. Already they are making their mark in the classroom, Basketball, Football & many other areas. I want to wish all our students & their parents a happy & peaceful mid-term. There will be many more activities and adventures over the coming months and of course a great deal of work Transition Year News Leaving Certificate Naughton Scholarships T.Y. Induction – Belfast Results Congratulations to Frances Activity Centre 29th August Wednesday 17th August was a Ryan & Fearghal Donaghy Congrats to Emma Heavin and very big day in the lives of last from Class of 2012 who were Ryan Callan who were chosen years 6thYears. It was marvel- the sole recipients of this pres- from over 350 students to take lous to see the delight and tigious award in Co. -

A Message from the Headmistress
A Message from the Headmistress Welcome to the 2012-13 edition concern for others and generosity of The Victorian magazine. At the are also expressed in charitable College, pupils learn, achieve and and community involvement. Our acquire the skills and values which potential for creativity in music, art will prepare them for adult life and and literature is showcased here in all for the world of work. This year’s its variety. magazine offers a record of the year inside and outside of the classroom 2012-13 was a very special year for and takes us from south Belfast to the sport in the College as our 1st XI continents of Europe, North America, Hockey Team played in the Schools’ Africa and Asia. Cup final for the first time in 40 years and brought us great pride. Sporting The magazine begins with a report life goes from strength to strength of the activities of the Association thanks to teams, individuals and all of Parents and Teachers which is those who teach, train and coach to always energetic in its support for whom we express our appreciation. the College, and we are very grateful to parents for their support and I thank Dr Mitchell-Barrett and the partnership, and likewise to our Board of Governors. Our staff of the Preparatory Department, Pre-School and Boarding Department, the House system, the contribution Playgroup for all their efforts to give our youngest pupils of pupils in the School Council and the visit of President such a welcome to the world of learning. I thank also and Mrs Obama are celebrated in words and pictures. -

Achieving Excellence Together
0 Achieving excellence together Achieving excellence together 1 Key Stage 3 Management Team FÁILTE Welcome to our Junior Presentation. On this occasion we mark the achievements of our Year 10 pupils, as we celebrate their attainment, both academic and extra-curricular. ORDER OF THE MORNING CLÁR NA MAIDINE 1. Mass 2. Welcome 3. Principal’s address 4. Presentation of certificates 5. Presentation of special awards 6. Invitation to refreshments 2 Achieving excellence together YEAR 10 MASS Theme: ‘Thanksgiving and Hope’ Entrance Hymn: We Praise You Greeting We have gathered here today to mark the end of Key Stage 3. It’s a time of joy and sadness. We celebrate the unity we have shared in the past year, the friends we have made, the fun we have had together and we ask God’s blessing on all our students and their families gathered here today. Penitential Act (All stand) I confess to almighty God and to you, my brothers and sisters, that I have greatly sinned, in my thoughts and in my words, in what I have done and in what I have failed to do, through my fault, through my fault, through my most grievous fault; therefore I ask blessed Mary ever-Virgin, all the Angels and Saints, and you, my brothers and sisters, to pray for me to the Lord our God. Opening Prayer We thank you, gracious God, for the gifts of life and of love; we thank you for the gifts of learning and of intelligence. We ask you that you will always help us to use these gifts in your service. -

Encyclopedia of CELTIC MYTHOLOGY and FOLKLORE
the encyclopedia of CELTIC MYTHOLOGY AND FOLKLORE Patricia Monaghan The Encyclopedia of Celtic Mythology and Folklore Copyright © 2004 by Patricia Monaghan All rights reserved. No part of this book may be reproduced or utilized in any form or by any means, electronic or mechanical, including photocopying, recording, or by any information storage or retrieval systems, without permission in writing from the publisher. For information contact: Facts On File, Inc. 132 West 31st Street New York NY 10001 Library of Congress Cataloging-in-Publication Data Monaghan, Patricia. The encyclopedia of Celtic mythology and folklore / Patricia Monaghan. p. cm. Includes bibliographical references and index. ISBN 0-8160-4524-0 (alk. paper) 1. Mythology, Celtic—Encyclopedias. 2. Celts—Folklore—Encyclopedias. 3. Legends—Europe—Encyclopedias. I. Title. BL900.M66 2003 299'.16—dc21 2003044944 Facts On File books are available at special discounts when purchased in bulk quantities for businesses, associations, institutions, or sales promotions. Please call our Special Sales Department in New York at (212) 967-8800 or (800) 322-8755. You can find Facts On File on the World Wide Web at http://www.factsonfile.com Text design by Erika K. Arroyo Cover design by Cathy Rincon Printed in the United States of America VB Hermitage 10 9 8 7 6 5 4 3 2 1 This book is printed on acid-free paper. CONTENTS 6 INTRODUCTION iv A TO Z ENTRIES 1 BIBLIOGRAPHY 479 INDEX 486 INTRODUCTION 6 Who Were the Celts? tribal names, used by other Europeans as a The terms Celt and Celtic seem familiar today— generic term for the whole people. -

Supreme Court Annual Report 2020
2020Annual Report Report published by the Supreme Court of Ireland with the support of the Courts Service An tSeirbhís Chúirteanna Courts Service Editors: Sarahrose Murphy, Senior Executive Legal Officer to the Chief Justice Patrick Conboy, Executive Legal Officer to the Chief Justice Case summaries prepared by the following Judicial Assistants: Aislinn McCann Seán Beatty Iseult Browne Senan Crawford Orlaith Cross Katie Cundelan Shane Finn Matthew Hanrahan Cormac Hickey Caoimhe Hunter-Blair Ciara McCarthy Rachael O’Byrne Mary O’Rourke Karl O’Reilly © Supreme Court of Ireland 2020 2020 Annual Report Table of Contents Foreword by the Chief Justice 6 Introduction by the Registrar of the Supreme Court 9 2020 at a glance 11 Part 1 About the Supreme Court of Ireland 15 Branches of Government in Ireland 16 Jurisdiction of the Supreme Court 17 Structure of the Courts of Ireland 19 Timeline of key events in the Supreme Court’s history 20 Seat of the Supreme Court 22 The Supreme Court Courtroom 24 Journey of a typical appeal 26 Members of the Supreme Court 30 The Role of the Chief Justice 35 Retirement and Appointments 39 The Constitution of Ireland 41 Depositary for Acts of the Oireachtas 45 Part 2 The Supreme Court in 2020 46 COVID-19 and the response of the Court 47 Remote hearings 47 Practice Direction SC21 48 Application for Leave panels 48 Statement of Case 48 Clarification request 48 Electronic delivery of judgments 49 Sitting in King’s Inns 49 Statistics 50 Applications for Leave to Appeal 50 Categorisation of Applications for Leave to Appeal -

Last Name First Name Grant Amount (£) Art Form Adams Sharon 2,150 Craft Anderson Hannah 2,500 Circus/Street Theatre/Carnival Ar
Last Name First Name Grant Amount (£) Art form Adams Sharon 2,150 Craft Anderson Hannah 2,500 Circus/Street Theatre/Carnival Arthur Kate Caoimhe 2,500 Literature Arthur Margaret 1,000 Visual Arts Athanasiou Chrysovalanti 1,000 Visual Arts Bennett Ed 2,500 Music Booth Adam 2,500 Music Bourke Kyron 1,500 Drama Bradley Stephen 1,000 Visual Arts Breen Laura 2,500 Craft Brown Katie 2,500 Craft Browne Jemma 1,980 Craft Burnside Connor 2,500 Music Butler Anne 2,500 Craft Cahoon Neal 1,000 Literature Caldwell Conor 2,500 Traditional Arts Cameron Martin 2,500 Craft Campbell Iain 1,000 Literature Cannon Diane 2,500 Traditional Arts Cargnelli Alessia 2,500 Visual Arts Carr Ruth 2,500 Literature Carson Julie Anne (Jan) 1,687 Literature Carson Paula 1,850 Drama Carten Mairead 2,500 Visual Arts Casey Nollaig 2,500 Traditional Arts Clarkson Amelia 800 Music Clements Paul 2,175 Literature Collins Christina 2,500 Literature Conaghan Ruairi 2,285 Drama Conn Stephanie 2,000 Literature Connell Benjamin 1,125 Architecture Cooper Emily 2,500 Literature Corry Ruth 2,500 Traditional Arts Courtney Roger 930 Literature Covington Wilhelmina Peace 1,000 Visual Arts Craig-McGrath Constance 1,900 Drama Creighton Kelly 2,000 Literature Crothers Ruth 2,500 Visual Arts Cuddington Natasha 2,500 Literature Cullinane Liz 2,500 Drama Cunningham Alan 1,500 Literature Cunningham Tanya 1,500 Craft Cunningham-Bell Sara 2,221 Visual Arts Cupples Conor 1,750 Drama Davies Eli 2,400 Literature Daye-Hutchinson Karen 2,500 Visual Arts Deery Aidan 2,500 Music Denvir Owen 2,500 Music -

Irish Sport Horse Stallions
Irish Sport Horse Stallions Irish Sport Horse Stallions An extract from the Irish Sport Horse Studbook Stallion Book The Irish Sport Horse Studbook is maintained by Horse Sport Ireland and the Northern Ireland Horse Board Horse Sport Ireland First Floor, Beech House, Millennium Park, Osberstown, Naas, Co. Kildare, Ireland Telephone: 045 850800. Int: +353 45 850800 Fax: 045 850850. Int: +353 45 850850 Email: [email protected] Website: www.horsesportireland.ie Northern Ireland Horse Board Office Suite, Meadows Equestrian Centre Embankment Road, Lurgan Co. Armagh, BT66 6NE, Northern Ireland Telephone: 028 38 343355 Fax: 028 38 325332 Email: [email protected] Website: www.nihorseboard.org Copyright © Horse Sport Ireland 2015 INDEX OF IRISH SPORT HORSE STALLIONS INDEX OF IRISH SPORT HORSE STALLIONS IRISH SPORT HORSE STALLIONS IRISH SPORT HORSE STALLIONS ARDCOLUM DUKE {TIH} .................. 4 FURISTO'S DIAMOND.................... 39 (Supplementary Approved) GALLANT CAVALIER....................... 40 BAHRAIN CRUISE {TIH} .................... 5 CHIPPISON .....................................14 BEOWULF ........................................ 6 GATSBY.......................................... 41 BOHERDEAL CLOVER {TIH} .............. 6 KENNEDYS CLOVER {TIH} .............. 43 IRISH SPORT HORSE STALLIONS CAPTAIN CARNUTE.......................... 7 KINGS MASTER {TIH}..................... 43 (Preliminary Approved) CAPTAIN CLOVER {TIH}.................... 8 KNOCK DIAMOND VIEW {TIH} ...... 45 CARA TOUCHE............................... 10 -

Gold for Harrington & Silver for Mulcaire
Gold For Harrington & Silver for Mulcaire 6 Munster athletes were selected for Ireland for the SIAB Schools Cross Country International on Saturday 22nd March in Bolton, England – Junior Girls Caoimhe Harrington, Colaiste Pobail, Bheanntrai Laura Nicholson, Bandon Grammer School Intermediate Girls Fiona Everard, Maria Immaculata CC, Dunmanway Intermediate Boys Kevin Mulcaire St Flannan’s College, Ennis, Co Clare Barry Keane, St. Declan’s, Kilmacthomas Fearghal Curtin, Midleton CBS Caoimhe Harrington, West Muskerry A.C. representing her school Colaiste Pobail, Bheanntrai put in a terrific performance to lead the field home in the Junior Girls race winning individual gold & leading the Irish team to a bronze medal in the team competition which included Laura Nicholson, Bandon Grammer School (Bandon A.C.) who finished 25th overall. Fiona Everard, Maria Immaculate CC Dunmanway (Bandon A.C.) won a bronze team medal with the Irish team in the Intermediate competition coming home in 22nd place overall. Kevin Mulcaire, Ennis Track A.C. representing his school, St. Flannan’s Ennis took silver in the Intermediate Boys race with a wonderful display of running & led his team home to a silver medal victory in the team competition which included fellow Munster men, Fearghal Curtin, Midleton CBS (Youghal A.C.) who was 13th overall & Barry Keane, St. Declan’s, Kilmacthomas in 17th place. Congratulations & well done to all 6 athletes. Full results at end of this report. See Extract from AAI Website dated Monday, 24th March below:- “There were thrilling individual victories achieved by two young Irish athletes; Caoimhe Harrington (Colaiste Pobail, Bheanntrai , Bantry) and Paddy Maher (Dunshaughlin Community College) at the SIAB International Schools Cross Country event in Bolton and there was silver and bronze medal success too by Kevin Mulcaire (St Flannan’s Ennis) and James Maguire (St Benildus, Dublin) in their respective Intermediate and Junior Boys events.