Integrated Stress Response of Vertebrates Is Regulated by Four
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CEP41 As an ASD Gene Ashok Patowary 1,Soyeonwon2,Shinjioh2,Ryanrnesbitt1, Marilyn Archer1, Debbie Nickerson3, Wendy H
Patowary et al. Translational Psychiatry (2019) 9:4 https://doi.org/10.1038/s41398-018-0343-z Translational Psychiatry ARTICLE Open Access Family-based exome sequencing and case- control analysis implicate CEP41 as an ASD gene Ashok Patowary 1,SoYeonWon2,ShinJiOh2,RyanRNesbitt1, Marilyn Archer1, Debbie Nickerson3, Wendy H. Raskind1,4, Raphael Bernier1,JiEunLee2,5 and Zoran Brkanac1 Abstract Autism Spectrum Disorder (ASD) is a complex neurodevelopmental disorder with a strong genetic component. Although next-generation sequencing (NGS) technologies have been successfully applied to gene identification in de novo ASD, the genetic architecture of familial ASD remains largely unexplored. Our approach, which leverages the high specificity and sensitivity of NGS technology, has focused on rare variants in familial autism. We used NGS exome sequencing in 26 families with distantly related affected individuals to identify genes with private gene disrupting and missense variants of interest (VOI). We found that the genes carrying VOIs were enriched for biological processes related to cell projection organization and neuron development, which is consistent with the neurodevelopmental hypothesis of ASD. For a subset of genes carrying VOIs, we then used targeted NGS sequencing and gene-based variant burden case-control analysis to test for association with ASD. Missense variants in one gene, CEP41, associated significantly with ASD (p = 6.185e−05). Homozygous gene-disrupting variants in CEP41 were initially found to be responsible for recessive Joubert syndrome. Using a zebrafish model, we evaluated the mechanism by which the 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; CEP41 variants might contribute to ASD. We found that CEP41 missense variants affect development of the axonal tract, cranial neural crest migration and social behavior phenotype. -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

De Novo EIF2AK1 and EIF2AK2 Variants Are Associated with Developmental Delay, Leukoencephalopathy, and Neurologic Decompensation
bioRxiv preprint doi: https://doi.org/10.1101/757039; this version posted September 16, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. De novo EIF2AK1 and EIF2AK2 variants are associated with developmental delay, leukoencephalopathy, and neurologic decompensation Dongxue Mao1,2, Chloe M. Reuter3,4, Maura R.Z. Ruzhnikov5,6, Anita E. Beck7, Emily G. Farrow8,9,10, Lisa T. Emrick1,11,12,13, Jill A. Rosenfeld12, Katherine M. Mackenzie5, Laurie Robak2,12,13, Matthew T. Wheeler3,14, Lindsay C. Burrage12,13, Mahim Jain15, Pengfei Liu12, Daniel Calame11,13, Sebastien Küry17,18, Martin Sillesen19, Klaus Schmitz-Abe20, Davide Tonduti21, Luigina Spaccini22, Maria Iascone23, Casie A. Genetti20, Madeline Graf16, Alyssa Tran12, Mercedes Alejandro12, Undiagnosed Diseases Network, Brendan H. Lee12,13, Isabelle Thiffault8,9,24, Pankaj B. Agrawal#,20, Jonathan A. Bernstein#,3,25, Hugo J. Bellen#,2,12,26,27,28, Hsiao- Tuan Chao#,1,2,11,12,13,28,27,29 #Correspondence should be addressed: [email protected] (P.A.), [email protected] (J.A.B.), [email protected] (H.J.B.), [email protected] (H.T.C.) 1Department of Pediatrics, Baylor College of Medicine (BCM), Houston, TX 2Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Houston, TX 3Stanford Center for Undiagnosed Diseases, Stanford University, Stanford, CA 4Stanford Center for Inherited Cardiovascular Disease, Division of Cardiovascular Medicine, -

Unique Integrated Stress Response Sensors Regulate Cancer Cell
RESEARCH ARTICLE Unique integrated stress response sensors regulate cancer cell susceptibility when Hsp70 activity is compromised Sara Sannino1*, Megan E Yates2,3,4, Mark E Schurdak5,6, Steffi Oesterreich2,3,7, Adrian V Lee2,3,7, Peter Wipf8, Jeffrey L Brodsky1* 1Department of Biological Sciences, University of Pittsburgh, Pittsburgh, United States; 2Women’s Cancer Research Center, UPMC Hillman Cancer Center, Magee- Women Research Institute, Pittsburgh, United States; 3Integrative Systems Biology Program, University of Pittsburgh, Pittsburgh, United States; 4Medical Scientist Training Program, University of Pittsburgh School of Medicine, Pittsburgh, United States; 5Department of Computational and Systems Biology, University of Pittsburgh, Pittsburgh, United States; 6University of Pittsburgh Drug Discovery Institute, Pittsburgh, United States; 7Department of Pharmacology and Chemical Biology, University of Pittsburgh School of Medicine, Pittsburgh, United States; 8Department of Chemistry, University of Pittsburgh, Pittsburgh, United States Abstract Molecular chaperones, such as Hsp70, prevent proteotoxicity and maintain homeostasis. This is perhaps most evident in cancer cells, which overexpress Hsp70 and thrive even when harboring high levels of misfolded proteins. To define the response to proteotoxic challenges, we examined adaptive responses in breast cancer cells in the presence of an Hsp70 inhibitor. We discovered that the cells bin into distinct classes based on inhibitor sensitivity. Strikingly, the most resistant cells have higher autophagy levels, and autophagy was maximally activated only in resistant cells upon Hsp70 inhibition. In turn, resistance to compromised Hsp70 *For correspondence: function required the integrated stress response transducer, GCN2, which is commonly associated [email protected] (SS); with amino acid starvation. In contrast, sensitive cells succumbed to Hsp70 inhibition by activating [email protected] (JLB) PERK. -

Relevance of Translation Initiation in Diffuse Glioma Biology and Its
cells Review Relevance of Translation Initiation in Diffuse Glioma Biology and its Therapeutic Potential Digregorio Marina 1, Lombard Arnaud 1,2, Lumapat Paul Noel 1, Scholtes Felix 1,2, Rogister Bernard 1,3 and Coppieters Natacha 1,* 1 Laboratory of Nervous System Disorders and Therapy, GIGA-Neurosciences Research Centre, University of Liège, 4000 Liège, Belgium; [email protected] (D.M.); [email protected] (L.A.); [email protected] (L.P.N.); [email protected] (S.F.); [email protected] (R.B.) 2 Department of Neurosurgery, CHU of Liège, 4000 Liège, Belgium 3 Department of Neurology, CHU of Liège, 4000 Liège, Belgium * Correspondence: [email protected] Received: 18 October 2019; Accepted: 26 November 2019; Published: 29 November 2019 Abstract: Cancer cells are continually exposed to environmental stressors forcing them to adapt their protein production to survive. The translational machinery can be recruited by malignant cells to synthesize proteins required to promote their survival, even in times of high physiological and pathological stress. This phenomenon has been described in several cancers including in gliomas. Abnormal regulation of translation has encouraged the development of new therapeutics targeting the protein synthesis pathway. This approach could be meaningful for glioma given the fact that the median survival following diagnosis of the highest grade of glioma remains short despite current therapy. The identification of new targets for the development of novel therapeutics is therefore needed in order to improve this devastating overall survival rate. This review discusses current literature on translation in gliomas with a focus on the initiation step covering both the cap-dependent and cap-independent modes of initiation. -

1 Kinobead Profiling Reveals Reprogramming of B-Cell Receptor Signaling in Response to Therapy Within Primary CLL Cells. Linley
bioRxiv preprint doi: https://doi.org/10.1101/841312; this version posted November 14, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Kinobead profiling reveals reprogramming of B-cell receptor signaling in response to therapy within primary CLL cells. Linley AJ1, Griffin R2, Cicconi S2, D’Avola A3$, MacEwan DJ4, Pettit AR1, Kalakonda N1, Packham G3, Prior IA5, Slupsky JR1. 1. Department of Molecular and Clinical Cancer Medicine, Institute of Translational Medicine, University of Liverpool, Sherrington Building, Ashton Street, Liverpool, UK. 2. CRUK Clinical Trials Unit, University of Liverpool, Waterhouse Building, Ashton Street, Liverpool. 3. Southampton Cancer Research UK Centre, Cancer Sciences Unit, Faculty of Medicine, University of Southampton, Southampton, UK. 4. Department of Molecular and Clinical Pharmacology, Institute of Translational Medicine, University of Liverpool, Sherrington Building, Ashton Street, Liverpool, UK. 5. Department of Cellular and Molecular Physiology, Institute of Translational Medicine, University of Liverpool, Nuffield Wing, Crown Street, Liverpool, UK. $ Current Address: The Francis Crick Institute, 1 Midland Road, London, UK. Corresponding author: Dr Adam J Linley, Department of Molecular and Clinical Cancer Medicine, Institute of Translational Medicine, University of Liverpool, Sherrington Building, Ashton Street, Liverpool, UK; [email protected]; +44(0)151 794 5310 Running head: Therapy brings about BCR signal changes. Key points 1. sIgM signaling patterns alter following in vivo therapy using either chemoimmunotherapy or ibrutinib. 2. Kinobeads provide a novel method for high-resolution investigation of signaling in primary CLL cells. 1 bioRxiv preprint doi: https://doi.org/10.1101/841312; this version posted November 14, 2019. -

Blueprint Genetics Congenital Structural Heart Disease Panel
Congenital Structural Heart Disease Panel Test code: CA1501 Is a 114 gene panel that includes assessment of non-coding variants. Is ideal for patients with congenital heart disease, particularly those with features of hereditary disorders. Is not ideal for patients suspected to have a ciliopathy or a rasopathy. For those patients, please consider our Primary Ciliary Dyskinesia Panel and our Noonan Syndrome Panel, respectively. About Congenital Structural Heart Disease There are many types of congenital heart disease (CHD) ranging from simple asymptomatic defects to complex defects with severe, life-threatening symptoms. CHDs are the most common type of birth defect and affect at least 8 out of every 1,000 newborns. Annually, more than 35,000 babies in the United States are born with CHDs. Many of these CHDs are simple conditions and need no treatment or are easily repaired. Some babies are born with complex CHD requiring special medical care. The diagnosis and treatment of complex CHDs has greatly improved over the past few decades. As a result, almost all children who have complex heart defects survive to adulthood and can live active, productive lives. However, many patients who have complex CHDs continue to need special heart care throughout their lives. In the United States, more than 1 million adults are living with congenital heart disease. Availability 4 weeks Gene Set Description Genes in the Congenital Structural Heart Disease Panel and their clinical significance Gene Associated phenotypes Inheritance ClinVar HGMD ABL1 Congenital -

PRODUCTS and SERVICES Target List
PRODUCTS AND SERVICES Target list Kinase Products P.1-11 Kinase Products Biochemical Assays P.12 "QuickScout Screening Assist™ Kits" Kinase Protein Assay Kits P.13 "QuickScout Custom Profiling & Panel Profiling Series" Targets P.14 "QuickScout Custom Profiling Series" Preincubation Targets Cell-Based Assays P.15 NanoBRET™ TE Intracellular Kinase Cell-Based Assay Service Targets P.16 Tyrosine Kinase Ba/F3 Cell-Based Assay Service Targets P.17 Kinase HEK293 Cell-Based Assay Service ~ClariCELL™ ~ Targets P.18 Detection of Protein-Protein Interactions ~ProbeX™~ Stable Cell Lines Crystallization Services P.19 FastLane™ Structures ~Premium~ P.20-21 FastLane™ Structures ~Standard~ Kinase Products For details of products, please see "PRODUCTS AND SERVICES" on page 1~3. Tyrosine Kinases Note: Please contact us for availability or further information. Information may be changed without notice. Expression Protein Kinase Tag Carna Product Name Catalog No. Construct Sequence Accession Number Tag Location System HIS ABL(ABL1) 08-001 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) ABL(ABL1) BTN BTN-ABL(ABL1) 08-401-20N Full-length 2-1130 NP_005148.2 N-terminal DYKDDDDK Insect (sf21) ABL(ABL1) [E255K] HIS ABL(ABL1)[E255K] 08-094 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) HIS ABL(ABL1)[T315I] 08-093 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) ABL(ABL1) [T315I] BTN BTN-ABL(ABL1)[T315I] 08-493-20N Full-length 2-1130 NP_005148.2 N-terminal DYKDDDDK Insect (sf21) ACK(TNK2) GST ACK(TNK2) 08-196 Catalytic domain -

Cardiovascular Diseases Genetic Testing Program Information
Cardiovascular Diseases Genetic Testing Program Description: Congenital Heart Disease Panels We offer comprehensive gene panels designed to • Congenital Heart Disease Panel (187 genes) diagnose the most common genetic causes of hereditary • Heterotaxy Panel (114 genes) cardiovascular diseases. Testing is available for congenital • RASopathy/Noonan Spectrum Disorders Panel heart malformation, cardiomyopathy, arrythmia, thoracic (31 genes) aortic aneurysm, pulmonary arterial hypertension, Marfan Other Panels syndrome, and RASopathy/Noonan spectrum disorders. • Pulmonary Arterial Hypertension (PAH) Panel Hereditary cardiovascular disease is caused by variants in (20 genes) many different genes, and may be inherited in an autosomal dominant, autosomal recessive, or X-linked manner. Other Indications: than condition-specific panels, we also offer single gene Panels: sequencing for any gene on the panels, targeted variant • Confirmation of genetic diagnosis in a patient with analysis, and targeted deletion/duplication analysis. a clinical diagnosis of cardiovascular disease Tests Offered: • Carrier or pre-symptomatic diagnosis identification Arrythmia Panels in individuals with a family history of cardiovascular • Comprehensive Arrhythmia Panel (81 genes) disease of unknown genetic basis • Atrial Fibrillation (A Fib) Panel (28 genes) Gene Specific Sequencing: • Atrioventricular Block (AV Block) Panel (7 genes) • Confirmation of genetic diagnosis in a patient with • Brugada Syndrome Panel (21 genes) cardiovascular disease and in whom a specific -

Brain-Restricted Mtor Inhibition with Binary Pharmacology 2 3 Ziyang Zhang1, Qiwen Fan2,3, Xujun Luo2,3, Kevin J
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.12.336677; this version posted October 12, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Brain-Restricted mTOR Inhibition with Binary Pharmacology 2 3 Ziyang Zhang1, Qiwen Fan2,3, Xujun Luo2,3, Kevin J. Lou1, William A. Weiss2,3,4,5, Kevan 4 M. Shokat1,* 5 6 1 Department of Cellular and Molecular Pharmacology and Howard Hughes Medical 7 Institute, University of California, San Francisco, California 8 2 Helen Diller Family Comprehensive Cancer Center, San Francisco, California 9 3 Department of Neurology, University of California, San Francisco, California 10 4 Department of Pediatrics, University of California, San Francisco, California 11 5 Department of Neurological Surgery, University of California, San Francisco, California 12 * Corresponding author. [email protected] (K.M.S.). 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.10.12.336677; this version posted October 12, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 13 Abstract 14 On-target-off-tissue drug engagement is an important source of adverse effects 15 that constrains the therapeutic window of drug candidates. In diseases of the central 16 nervous system, drugs with brain-restricted pharmacology are highly desirable. -

Identification of EMT-Related High-Risk Stage II Colorectal Cancer And
www.nature.com/bjc ARTICLE Molecular Diagnostics Identification of EMT-related high-risk stage II colorectal cancer and characterisation of metastasis-related genes Kai Wang1, Kai Song1, Zhigang Ma2, Yang Yao2, Chao Liu2, Jing Yang1, Huiting Xiao1, Jiashuai Zhang1, Yanqiao Zhang2 and Wenyuan Zhao1 BACKGROUND: Our laboratory previously reported an individual-level prognostic signature for patients with stage II colorectal cancer (CRC). However, this signature was not applicable for RNA-sequencing datasets. In this study, we constructed a robust epithelial-to-mesenchymal transition (EMT)- related gene pair prognostic signature. METHODS: Based on EMT-related genes, metastasis-associated gene pairs were identified between metastatic and non-metastatic samples. Then, we selected prognosis-associated gene pairs, which were significantly correlated with disease-free survival of stage II CRC using multivariate Cox regression model, as the EMT-related prognosis signature. RESULTS: An EMT-related signature composed of fifty-one gene pairs (51-GPS) for prediction-relapse risk of patients with stage II CRC was developed, whose prognostic efficiency was validated in independent datasets. Moreover, 51-GPS achieved better predictive performance than other reported signatures, including a commercial signature Oncotype Dx colon cancer and an immune-related gene pair signature. Besides, EMT-related functional gene sets achieved high enrichment scores in high-risk samples. Especially, loss-of-function antisense approach showed that DEGs between the predicted two clusters were metastasis- related. CONCLUSIONS: The EMT-related gene pair signature can identify the high relapse-risk patients with stage II CRC, which can facilitate individualised management of patients. British Journal of Cancer (2020) 123:410–417; https://doi.org/10.1038/s41416-020-0902-y BACKGROUND greatly affected by the sampling locations10,11 and RNA degrada- Colorectal cancer (CRC) ranks third in terms of incidence, but tion problem during sample preparation12 of tumour tissues. -
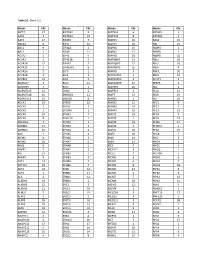
Table S1. Gene List Genes Chr Genes Chr Genes Chr Genes Chr SEPT9
Table S1. Gene List Genes Chr Genes Chr Genes Chr Genes Chr SEPT9 17 EIF2AK3 2 MAPK14 6 RASSF5 1 AAK1 2 EIF2AK4 15 MAPK15 8 RAVER2 1 AATK 17 EIF2B5 3 MAPK3 16 RAX2 19 ABCA1 9 EIF3J 15 MAPK4 18 RB1 13 ABL1 9 EIF4A2 3 MAPK6 15 RBBP4 1 ABL2 1 EIF4B 12 MAPK7 17 RBBP5 1 ACTR2 2 EIF4E 4 MAPK8 10 RBBP8 18 ACVR1 2 EIF4E1B 5 MAPK8IP1 11 RBL1 20 ACVR1B 12 EIF4E2 2 MAPK8IP2 22 RBL2 16 ACVR1C 2 EIF4EBP1 8 MAPK8IP3 16 RBPJ 4 ACVR2A 2 ELF3 1 MAPK9 5 RBPJL 20 ACVR2B 3 ELF4 X MAPKAPK2 1 RBX1 22 ACVRL1 12 ELK1 X MAPKAPK3 3 RCHY1 4 ADAM17 2 ELK3 12 MAPKAPK5 12 REEP5 5 ADAM29 4 ELK4 1 MAPRE1 20 REL 2 ADAMTS15 11 EML4 2 MAPRE3 2 RELA 11 ADAMTS18 16 ENDOD1 11 MAPT 17 RELB 19 ADAMTSL3 15 ENDOG 9 MARK1 1 RET 10 ADCK1 14 EP300 22 MARK2 11 RFC1 4 ADCK2 7 EPAS1 2 MARK3 14 RFC2 7 ADCK3 1 EPCAM 2 MARK4 19 RFC3 13 ADCK4 19 EPHA1 7 MAST1 19 RFC4 3 ADCK5 8 EPHA10 1 MAST2 1 RFC5 12 ADORA1 1 EPHA2 1 MAST3 19 RFNG 17 ADRBK1 11 EPHA3 3 MAST4 5 RFWD2 1 ADRBK2 22 EPHA4 2 MASTL 10 RFX2 19 AGK 7 EPHA5 4 MATK 19 RHEB 7 AIFM1 X EPHA6 3 MAX 14 RHO 3 AIFM2 10 EPHA7 6 MBIP 14 RHOA 3 AIM1 6 EPHA8 1 MCC 5 RHOC 1 AIMP2 7 EPHB1 3 MCF2L2 3 RHOQ 2 AIP 11 EPHB2 1 MCL1 1 RICTOR 5 AKAP4 X EPHB3 3 MCM2 3 RIOK1 6 AKT1 14 EPHB4 7 MCM3 6 RIOK2 5 AKT1S1 19 EPHB6 7 MCM4 8 RIOK3 18 AKT2 19 EPS8 12 MCM5 22 RIPK1 6 AKT3 1 ERBB2 17 MCM6 2 RIPK2 8 ALK 2 ERBB3 12 MCM7 7 RIPK3 14 ALKBH1 14 ERBB4 2 MCM8 20 RIPK4 21 ALKBH2 12 ERC2 3 MDM2 12 RMI1 9 ALKBH3 11 ERCC1 19 MDM4 1 RNASEL 1 ALMS1 2 ERCC2 19 MECOM 3 RNF213 17 ALPK1 4 ERCC3 2 MED12 X RNF220 1 ALPK2 18 ERCC4 16 MED12L 3 ROCK1 18 ALPK3 15 ERCC5 13