(12) Patent Application Publication (10) Pub. No.: US 2004/0067506 A1 Scheres Et Al
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

SEED LEAFLET No
SEED LEAFLET No. 84 September 2003 Baikiaea plurijuga Harms Taxonomy and nomenclature Botanical description Family: Fabaceae (Caesalpinioideae) A medium to large tree, 8-15 (20) m tall, with a large, Synonyms: none. dense, spreading crown. The bark is smooth and pale Vernacular/common names: Rhodesian teak, Zambe- at first, on older trees becoming fissured and cracked. zi teak, Zambezi redwood, Zimbabwean teak, Zambian Leaves are alternate and compound with 4 to 5 pairs teak (Eng.); mukusi (Botswana and Zambia); Rhode- of opposite leaflets. Each leaflet is up to 7 cm long, siese kiaat (South Africa); Zimbabwean teak, Zimbab- sparingly hairy especially on the lower surface and wean chestnut, umgusi, mukusi (Zimbabwe). midrib; the tip is rounded. The large, pink flowers are very attractive; they are borne in up to 30 cm long Distribution and habitat inflorescences. The species is confined to lowland tropical forests on the deep Kalahari sands between 13 and 20°S. It oc- curs naturally in Angola, Botswana, Namibia, Zambia and Zimbabwe, in areas with annual rainfall of 600- 1000 mm and a dry season of 6-8 months. Mature trees can withstand extreme temperatures of over 40°C and have been known to survive severe frost down to -15°C. It is mainly found in deep, infertile, sandy soils where it survives by developing a deep tap root. During the last century, most of the original Zambesi teak forests have been heavily exploited by logging, clearing of land for agriculture and frequent fires and the species is now mainly found in open, dry, deciduous woodland. The species is most typically associated with Pterocarpus angolensis, Julbernardia paniculata, Dialium englerianum. -

Investigating the Impact of Fire on the Natural Regeneration of Woody Species in Dry and Wet Miombo Woodland
Investigating the impact of fire on the natural regeneration of woody species in dry and wet Miombo woodland by Paul Mwansa Thesis presented in fulfilment of the requirements for the degree of Master of Science of Forestry and Natural Resource Science in the Faculty of AgriSciences at Stellenbosch University Supervisor: Prof Ben du Toit Co-supervisor: Dr Vera De Cauwer March 2018 Stellenbosch University https://scholar.sun.ac.za Declaration By submitting this thesis electronically, I declare that the entirety of the work contained therein is my own, original work, that I am the sole author thereof (save to the extent explicitly otherwise stated), that reproduction and publication thereof by Stellenbosch University will not infringe any third party rights and that I have not previously in its entirety or in part submitted it for obtaining any qualification. March 2018 Copyright © 2018 Stellenbosch University All rights reserved i Stellenbosch University https://scholar.sun.ac.za Abstract The miombo woodland is an extensive tropical seasonal woodland and dry forest formation in extent of 2.7 million km². The woodland contributes highly to maintenance and improvement of people’s livelihood security and stable growth of national economies. The woodland faces a wide range of disturbances including fire that affect vegetation structure. An investigation into the impact of fire on the natural regeneration of six tree species was conducted along a rainfall gradient. Baikiaea plurijuga, Burkea africana, Guibourtia coleosperma, Pterocarpus angolensis, Schinziophyton rautanenii and Terminalia sericea were selected on basis of being an important timber and/or utilitarian species, and the assumed abundance. The objectives of the study were to examine floristic composition, density and composition of natural regeneration; stand structure and vegetation cover within recently burnt (RB) and recently unburnt (RU) sections of the forest. -

Miombo Ecoregion Vision Report
MIOMBO ECOREGION VISION REPORT Jonathan Timberlake & Emmanuel Chidumayo December 2001 (published 2011) Occasional Publications in Biodiversity No. 20 WWF - SARPO MIOMBO ECOREGION VISION REPORT 2001 (revised August 2011) by Jonathan Timberlake & Emmanuel Chidumayo Occasional Publications in Biodiversity No. 20 Biodiversity Foundation for Africa P.O. Box FM730, Famona, Bulawayo, Zimbabwe PREFACE The Miombo Ecoregion Vision Report was commissioned in 2001 by the Southern Africa Regional Programme Office of the World Wide Fund for Nature (WWF SARPO). It represented the culmination of an ecoregion reconnaissance process led by Bruce Byers (see Byers 2001a, 2001b), followed by an ecoregion-scale mapping process of taxa and areas of interest or importance for various ecological and bio-physical parameters. The report was then used as a basis for more detailed discussions during a series of national workshops held across the region in the early part of 2002. The main purpose of the reconnaissance and visioning process was to initially outline the bio-physical extent and properties of the so-called Miombo Ecoregion (in practice, a collection of smaller previously described ecoregions), to identify the main areas of potential conservation interest and to identify appropriate activities and areas for conservation action. The outline and some features of the Miombo Ecoregion (later termed the Miombo– Mopane Ecoregion by Conservation International, or the Miombo–Mopane Woodlands and Grasslands) are often mentioned (e.g. Burgess et al. 2004). However, apart from two booklets (WWF SARPO 2001, 2003), few details or justifications are publically available, although a modified outline can be found in Frost, Timberlake & Chidumayo (2002). Over the years numerous requests have been made to use and refer to the original document and maps, which had only very restricted distribution. -

Biodiversity in Sub-Saharan Africa and Its Islands Conservation, Management and Sustainable Use
Biodiversity in Sub-Saharan Africa and its Islands Conservation, Management and Sustainable Use Occasional Papers of the IUCN Species Survival Commission No. 6 IUCN - The World Conservation Union IUCN Species Survival Commission Role of the SSC The Species Survival Commission (SSC) is IUCN's primary source of the 4. To provide advice, information, and expertise to the Secretariat of the scientific and technical information required for the maintenance of biologi- Convention on International Trade in Endangered Species of Wild Fauna cal diversity through the conservation of endangered and vulnerable species and Flora (CITES) and other international agreements affecting conser- of fauna and flora, whilst recommending and promoting measures for their vation of species or biological diversity. conservation, and for the management of other species of conservation con- cern. Its objective is to mobilize action to prevent the extinction of species, 5. To carry out specific tasks on behalf of the Union, including: sub-species and discrete populations of fauna and flora, thereby not only maintaining biological diversity but improving the status of endangered and • coordination of a programme of activities for the conservation of bio- vulnerable species. logical diversity within the framework of the IUCN Conservation Programme. Objectives of the SSC • promotion of the maintenance of biological diversity by monitoring 1. To participate in the further development, promotion and implementation the status of species and populations of conservation concern. of the World Conservation Strategy; to advise on the development of IUCN's Conservation Programme; to support the implementation of the • development and review of conservation action plans and priorities Programme' and to assist in the development, screening, and monitoring for species and their populations. -

The Australian Centre for International Agricultural Research (ACIAR) Was Established in June 1982 by an Act of the Australian Parliament
f The Australian Centre for International Agricultural Research (ACIAR) was established in June 1982 by an Act of the Australian Parliament. Its mandate is to help identify agricultural problems in developing countries and to commission collaborative research between Aus tralian and developing country researchers in fields where Australia has a special research competence. Where trade names are used this does not constitute endorsement of nor discrimination against any product by the Centre. ACIAR PROCEEDINGS This series of publications includes the full proceedings of research workshops or symposia organised or supported by ACIAR. Numbers in this series are distributed internationally to selected individuals and scientific institutions. Previous numbers in the series are listed on the inside back cover. " Australian Centre for International Agricultural Research G.P.O. Box 1571. Canberra, ACT 2601 Turnbull, J. W. 1990.Tropical tree seed research: proceedings of an international workshop held at the Forestry Training Centre, Gympie, Qld, Australia, 21-24 August 1989. ACIAR Proceedings No. 28, 156 p. ISBN 186320 004 5 Technical Editing: Janet Lawrence Typeset and laid out by: Abb-typesetting Pty Ltd, Collingwood, Vic. Printed by: Brown Prior Anderson Pty Ltd, Burwood, Vie. Tropical Tree Seed Research Proceedings of an international workshop held at the Forestry Training Centre, Gympie, Qld, Australia, 21-24 August 1989 Editor: J.W. Turnbull Host: International Union of Forest Research Organisations (IUFRO) Seed Problems Working Group Cosponsors: Australian International Development Assistance Bureau Australian Centre for International Agricultural Research CSIRO Division of Forestry and Forest Products Queensland Department of Forestry Organising Committee: Chairman: Stephen J. Midgley, CSIRO Division of Forestry and Forest Products Members: John C. -

Appendix 1 Vernacular Names
Appendix 1 Vernacular Names The vernacular names listed below have been collected from the literature. Few have phonetic spellings. Spelling is not helped by the difficulties of transcribing unwritten languages into European syllables and Roman script. Some languages have several names for the same species. Further complications arise from the various dialects and corruptions within a language, and use of names borrowed from other languages. Where the people are bilingual the person recording the name may fail to check which language it comes from. For example, in northern Sahel where Arabic is the lingua franca, the recorded names, supposedly Arabic, include a number from local languages. Sometimes the same name may be used for several species. For example, kiri is the Susu name for both Adansonia digitata and Drypetes afzelii. There is nothing unusual about such complications. For example, Grigson (1955) cites 52 English synonyms for the common dandelion (Taraxacum officinale) in the British Isles, and also mentions several examples of the same vernacular name applying to different species. Even Theophrastus in c. 300 BC complained that there were three plants called strykhnos, which were edible, soporific or hallucinogenic (Hort 1916). Languages and history are linked and it is hoped that understanding how lan- guages spread will lead to the discovery of the historical origins of some of the vernacular names for the baobab. The classification followed here is that of Gordon (2005) updated and edited by Blench (2005, personal communication). Alternative family names are shown in square brackets, dialects in parenthesis. Superscript Arabic numbers refer to references to the vernacular names; Roman numbers refer to further information in Section 4. -

SABONET Report No 18
ii Quick Guide This book is divided into two sections: the first part provides descriptions of some common trees and shrubs of Botswana, and the second is the complete checklist. The scientific names of the families, genera, and species are arranged alphabetically. Vernacular names are also arranged alphabetically, starting with Setswana and followed by English. Setswana names are separated by a semi-colon from English names. A glossary at the end of the book defines botanical terms used in the text. Species that are listed in the Red Data List for Botswana are indicated by an ® preceding the name. The letters N, SW, and SE indicate the distribution of the species within Botswana according to the Flora zambesiaca geographical regions. Flora zambesiaca regions used in the checklist. Administrative District FZ geographical region Central District SE & N Chobe District N Ghanzi District SW Kgalagadi District SW Kgatleng District SE Kweneng District SW & SE Ngamiland District N North East District N South East District SE Southern District SW & SE N CHOBE DISTRICT NGAMILAND DISTRICT ZIMBABWE NAMIBIA NORTH EAST DISTRICT CENTRAL DISTRICT GHANZI DISTRICT KWENENG DISTRICT KGATLENG KGALAGADI DISTRICT DISTRICT SOUTHERN SOUTH EAST DISTRICT DISTRICT SOUTH AFRICA 0 Kilometres 400 i ii Trees of Botswana: names and distribution Moffat P. Setshogo & Fanie Venter iii Recommended citation format SETSHOGO, M.P. & VENTER, F. 2003. Trees of Botswana: names and distribution. Southern African Botanical Diversity Network Report No. 18. Pretoria. Produced by University of Botswana Herbarium Private Bag UB00704 Gaborone Tel: (267) 355 2602 Fax: (267) 318 5097 E-mail: [email protected] Published by Southern African Botanical Diversity Network (SABONET), c/o National Botanical Institute, Private Bag X101, 0001 Pretoria and University of Botswana Herbarium, Private Bag UB00704, Gaborone. -

Chobe District Integrated Land Use Plan
Wageningen Environmental Research The mission of Wageningen University and Research is “To explore the potential P.O. Box 47 of nature to improve the quality of life”. Under the banner Wageningen University Chobe District Integrated Land Use Plan 6700 AB Wageningen & Research, Wageningen University and the specialised research institutes of The Netherlands the Wageningen Research Foundation have joined forces in contributing to T +31 (0) 317 48 07 00 inding solutions to important questions in the domain of healthy food and living www.wur.eu/environmental-research environment. With its roughly 30 branches, 5,000 employees and 10,000 students, Wageningen University & Research is one of the leading organisations in its domain. Report 2813 The unique Wageningen approach lies in its integrated approach to issues and ISSN 1566-7197 the collaboration between diff erent disciplines. Theo van der Sluis, Lin Cassidy, Chris Brooks, Piotr Wolski, Cornelis VanderPost, Piet Wit, Rene Henkens, Michiel van Eupen, Keta Mosepele, Oggie Maruapula, Elmar Veenendaal Chobe District Integrated Land Use Plan Theo van der Sluis1, Lin Cassidy2, Chris Brooks2, Piotr Wolski2, Cornelis VanderPost2, Piet Wit2, Rene Henkens1, Michiel van Eupen1, Keta Mosepele4, Oggie Maruapula2, Elmar Veenendaal3 1 Wageningen Environmental Research (Alterra) 2 Independent consultant 3 Wageningen University & Research 4 ORI This research is funded by UNDP, Contract nr. PR005/2016. Wageningen Environmental Research Wageningen, July 2017 Report 2813 ISSN 1566-7197 Theo van der Sluis, Lin Cassidy, Chris Brooks, Piotr Wolski, Cornelis VanderPost, Piet Wit, Rene Henkens, Michiel van Eupen, Keta Mosepele, Oggie Maruapula, Elmar Veenendaal, 2017. Chobe District Integrated Land Use Plan. Wageningen, Wageningen Environmental Research, Report 2813. -

Okavango Basin - Vegetation
Okavango Basin - Vegetation The vegetation of the Okavango basin is evergreen tree species Cryptosepalum their leaves for a period of some months shaped by a strong environmental gradient. exfoliatum ssp. pseudotaxus with its during the dry season. The dominance of From the northwest to the southeast mean microphyllous, compound leaves the canopy species depends on local abiotic annual precipitation diminishes strongly dominates extensive areas mostly on poor conditions and changes from Brachystegia and elevation drops from 1,850 m a.s.l. to ferralitic soils. It has been cited to be spiciformis to Julbernardia paniculata just 940 m a.s.l. in the Delta. In the intolerant of fire (Timberlake & together with various other woody species highlands of the Bié Plateau in central Chidumayo 2011) and hence is often (Fig. 4). Angola the topographic positions and associated with spares undergrowth but Miombo forests dominated by geologic substrate have a strong impact on thick moss layers and lianas. The evergreen Julbernardia paniculata: This vegetation vegetation patterns. However, the canopy creates rather moderate and humid unit covers the drier, southernmost belt of landscape in the middle reaches of the basin microclimatic habitat conditions and thus the Angolan Miombo woodlands and is rather flat and dominated by Kalahari facilitates the occurrence of species related represents the southern limit of Miombo sands; species composition here is mostly to central African forests (Fig. 1, 2 and 3). sensus stricto. Altitudes in this vegetation determined by nutrient availability or soil Miombo forests dominated by zone are considerably lower and hence water regime. In the Delta area topographic deciduous tree species: This type of these areas receive less precipitation. -

Biodiversity & Ecology
© University of Hamburg 2018 All rights reserved Klaus Hess Publishers Göttingen & Windhoek www.k-hess-verlag.de ISBN: 978-3-933117-95-3 (Germany), 978-99916-57-43-1 (Namibia) Language editing: Will Simonson (Cambridge), and Proofreading Pal Translation of abstracts to Portuguese: Ana Filipa Guerra Silva Gomes da Piedade Page desing & layout: Marit Arnold, Klaus A. Hess, Ria Henning-Lohmann Cover photographs: front: Thunderstorm approaching a village on the Angolan Central Plateau (Rasmus Revermann) back: Fire in the miombo woodlands, Zambia (David Parduhn) Cover Design: Ria Henning-Lohmann ISSN 1613-9801 Printed in Germany Suggestion for citations: Volume: Revermann, R., Krewenka, K.M., Schmiedel, U., Olwoch, J.M., Helmschrot, J. & Jürgens, N. (eds.) (2018) Climate change and adaptive land management in southern Africa – assessments, changes, challenges, and solutions. Biodiversity & Ecology, 6, Klaus Hess Publishers, Göttingen & Windhoek. Articles (example): Archer, E., Engelbrecht, F., Hänsler, A., Landman, W., Tadross, M. & Helmschrot, J. (2018) Seasonal prediction and regional climate projections for southern Africa. In: Climate change and adaptive land management in southern Africa – assessments, changes, challenges, and solutions (ed. by Revermann, R., Krewenka, K.M., Schmiedel, U., Olwoch, J.M., Helmschrot, J. & Jürgens, N.), pp. 14–21, Biodiversity & Ecology, 6, Klaus Hess Publishers, Göttingen & Windhoek. Corrections brought to our attention will be published at the following location: http://www.biodiversity-plants.de/biodivers_ecol/biodivers_ecol.php Biodiversity & Ecology Journal of the Division Biodiversity, Evolution and Ecology of Plants, Institute for Plant Science and Microbiology, University of Hamburg Volume 6: Climate change and adaptive land management in southern Africa Assessments, changes, challenges, and solutions Edited by Rasmus Revermann1, Kristin M. -
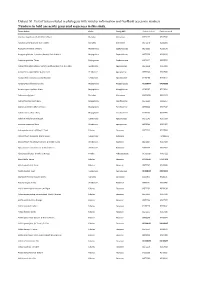
Dataset S1. List of Taxa Included in Phylogeny with Voucher Information and Genbank Accession Numbers
Dataset S1. List of taxa included in phylogeny with voucher information and GenBank accession numbers. Numbers in bold are newly generated sequences in this study. Taxon Author Order Family APG Genbank rbcLa Genbank matK Abutilon angulatum (Guill. & Perr.) Mast. Malvales Malvaceae JX572177 JX517944 Abutilon sonneratianum (Cav.) Sweet Malvales Malvaceae JX572178 JX518201 Acalypha chirindica S.Moore Malpighiales Euphorbiaceae JX572236 JX518178 Acalypha glabrata f. pilosior (Kuntze) Prain & Hutch. Malpighiales Euphorbiaceae JX572238 JX518120 Acalypha glabrata Thunb. Malpighiales Euphorbiaceae JX572237 JX517655 Acokanthera oblongifolia (Hochst.) Benth. & Hook.f. ex B.D.Jacks. Gentianales Apocynaceae JX572239 JX517911 Acokanthera oppositifolia (Lam.) Codd Gentianales Apocynaceae JX572240 JX517680 Acokanthera rotundata (Codd) Kupicha Gentianales Apocynaceae JF265266 JF270623 Acridocarpus chloropterus Oliv. Malpighiales Malpighiaceae KU568077 KX146299 Acridocarpus natalitius A.Juss. Malpighiales Malpighiaceae JF265267 JF270624 Adansonia digitata L. Malvales Malvaceae JQ025018 JQ024933 Adenia fruticosa Burtt Davy Malpighiales Passifloraceae JX572241 JX905957 Adenia gummifera (Harv.) Harms Malpighiales Passifloraceae JX572242 JX517347 Adenia spinosa Burtt Davy Malpighiales Passifloraceae JF265269 JX905950 Adenium multiflorum Klotzsch Gentianales Apocynaceae JX572243 JX517509 Adenium swazicum Stapf Gentianales Apocynaceae JX572244 JX517457 Adenopodia spicata (E.Mey.) C.Presl Fabales Fabaceae JX572245 JX517808 Afrocanthium lactescens (Hiern) Lantz -

Vegetation, Mammals, Birds, Reptiles, Fish, Insects, Aquatic Invertebrates and Ecosystems
AWF FOUR CORNERS TBNRM PROJECT : REVIEWS OF EXISTING BIODIVERSITY INFORMATION i Published for The African Wildlife Foundation's FOUR CORNERS TBNRM PROJECT by THE ZAMBEZI SOCIETY and THE BIODIVERSITY FOUNDATION FOR AFRICA 2004 PARTNERS IN BIODIVERSITY The Zambezi Society The Biodiversity Foundation for Africa P O Box HG774 P O Box FM730 Highlands Famona Harare Bulawayo Zimbabwe Zimbabwe Tel: +263 4 747002-5 E-mail: [email protected] E-mail: [email protected] Website: www.biodiversityfoundation.org Website : www.zamsoc.org The Zambezi Society and The Biodiversity Foundation for Africa are working as partners within the African Wildlife Foundation's Four Corners TBNRM project. The Biodiversity Foundation for Africa is responsible for acquiring technical information on the biodiversity of the project area. The Zambezi Society will be interpreting this information into user-friendly formats for stakeholders in the Four Corners area, and then disseminating it to these stakeholders. THE BIODIVERSITY FOUNDATION FOR AFRICA (BFA is a non-profit making Trust, formed in Bulawayo in 1992 by a group of concerned scientists and environmentalists. Individual BFA members have expertise in biological groups including plants, vegetation, mammals, birds, reptiles, fish, insects, aquatic invertebrates and ecosystems. The major objective of the BFA is to undertake biological research into the biodiversity of sub-Saharan Africa, and to make the resulting information more accessible. Towards this end it provides technical, ecological and biosystematic expertise. THE ZAMBEZI SOCIETY was established in 1982. Its goals include the conservation of biological diversity and wilderness in the Zambezi Basin through the application of sustainable, scientifically sound natural resource management strategies.