Effect of Salt Stress on the Expression and Promoter Methylation of the Genes Encoding the Mitochondrial and Cytosolic Forms of Aconitase and Fumarase in Maize
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Tbamitchodral L Alizaion of the 4Aminobutyrate-2-&Oxoglutarate
5d.em. J. (lWg77) 161,9O.-307 3O1 Printed in Great Britain Tbamitchodral L alizaion of the 4Aminobutyrate-2-&Oxoglutarate Transminase from Ox Brait By INGER SCHOUSDOE,* BIRGIT 1MO* and ARNE SCHOUSBOEt Department ofBDahemistry At andC*, University ofCopenhagen, 2200 Copenhagen M, Denark (Receved 4 June 1976) In order to determine the intramitochondrial location of 4-aminobutyrate transaminase, mitochondria were prepared from ox brain and freed from myelin and syiaptosomes by using conventional demitygradient-centrifugation techniques, and the purity was checked electron-microscopically. Iner and outer mimbrenes and matrix were prepared from the mitochondria by large-amplitude sweling and subsequent density-gradient centrfugationt The fractions were characterized by using both electron microscopy and differnt marker enzymes. From the specific activity of the 4-aminobutyrate transaminase in the submitochondrial fractions it was concluded that this enzyme is associated with the inner mitochondrial membrane. It is generally agreed that the 4-aminobutyrate-2- pyridoxal phosphate were from Sigma Chemical oxoglutarate transaminase (EC2.6.1.19) from brain is Co., St. Louis, MO, U.S.A. Ficoll was from mainly associated with free mitochondria (Salganicoff Pharmacia, Uppsala, Sweden, and crystallized & De Robertis, 1963, 1965; van den Berget al., 1965; bovine serum albumin was from BDH Biochemicals, van Kempen et at., 1965; Balazs et al., 1966; Poole, Dorset, U.K. 4-Amino[1-'4C]butyrate (sp. Waksman et al., 1968; Reijnierse et al., 1975), radioactivity 50mCi/mmol) and [1-14qtyramine (sp. and a preparation of a crude mitochondrial fraction radioactivity 9mCi/mmol) were obtained from was used by Schousboe et al. (1973) and Maitre et al. -

Alternative Acetate Production Pathways in Chlamydomonas Reinhardtii During Dark Anoxia and the Dominant Role of Chloroplasts in Fermentative Acetate Productionw
This article is a Plant Cell Advance Online Publication. The date of its first appearance online is the official date of publication. The article has been edited and the authors have corrected proofs, but minor changes could be made before the final version is published. Posting this version online reduces the time to publication by several weeks. Alternative Acetate Production Pathways in Chlamydomonas reinhardtii during Dark Anoxia and the Dominant Role of Chloroplasts in Fermentative Acetate ProductionW Wenqiang Yang,a,1 Claudia Catalanotti,a Sarah D’Adamo,b Tyler M. Wittkopp,a,c Cheryl J. Ingram-Smith,d Luke Mackinder,a Tarryn E. Miller,b Adam L. Heuberger,e Graham Peers,f Kerry S. Smith,d Martin C. Jonikas,a Arthur R. Grossman,a and Matthew C. Posewitzb a Carnegie Institution for Science, Department of Plant Biology, Stanford, California 94305 b Colorado School of Mines, Department of Chemistry and Geochemistry, Golden, Colorado 80401 c Stanford University, Department of Biology, Stanford, California 94305 d Clemson University, Department of Genetics and Biochemistry, Clemson, South Carolina 29634 e Colorado State University, Proteomics and Metabolomics Facility, Fort Collins, Colorado 80523 f Colorado State University, Department of Biology, Fort Collins, Colorado 80523 ORCID ID: 0000-0001-5600-4076 (W.Y.) Chlamydomonas reinhardtii insertion mutants disrupted for genes encoding acetate kinases (EC 2.7.2.1) (ACK1 and ACK2) and a phosphate acetyltransferase (EC 2.3.1.8) (PAT2, but not PAT1) were isolated to characterize fermentative acetate production. ACK1 and PAT2 were localized to chloroplasts, while ACK2 and PAT1 were shown to be in mitochondria. -

Alternative Oxidase: a Mitochondrial Respiratory Pathway to Maintain Metabolic and Signaling Homeostasis During Abiotic and Biotic Stress in Plants
Int. J. Mol. Sci. 2013, 14, 6805-6847; doi:10.3390/ijms14046805 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Review Alternative Oxidase: A Mitochondrial Respiratory Pathway to Maintain Metabolic and Signaling Homeostasis during Abiotic and Biotic Stress in Plants Greg C. Vanlerberghe Department of Biological Sciences and Department of Cell and Systems Biology, University of Toronto Scarborough, 1265 Military Trail, Toronto, ON, M1C1A4, Canada; E-Mail: [email protected]; Tel.: +1-416-208-2742; Fax: +1-416-287-7676 Received: 16 February 2013; in revised form: 8 March 2013 / Accepted: 12 March 2013 / Published: 26 March 2013 Abstract: Alternative oxidase (AOX) is a non-energy conserving terminal oxidase in the plant mitochondrial electron transport chain. While respiratory carbon oxidation pathways, electron transport, and ATP turnover are tightly coupled processes, AOX provides a means to relax this coupling, thus providing a degree of metabolic homeostasis to carbon and energy metabolism. Beside their role in primary metabolism, plant mitochondria also act as “signaling organelles”, able to influence processes such as nuclear gene expression. AOX activity can control the level of potential mitochondrial signaling molecules such as superoxide, nitric oxide and important redox couples. In this way, AOX also provides a degree of signaling homeostasis to the organelle. Evidence suggests that AOX function in metabolic and signaling homeostasis is particularly important during stress. These include abiotic stresses such as low temperature, drought, and nutrient deficiency, as well as biotic stresses such as bacterial infection. This review provides an introduction to the genetic and biochemical control of AOX respiration, as well as providing generalized examples of how AOX activity can provide metabolic and signaling homeostasis. -

Class-I and Class-II Fumarases Are a Paradigm of the Recruitment Of
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.04.232652; this version posted August 4, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Class-I and Class-II fumarases are a paradigm of the recruitment of 2 metabolites and metabolic enzymes for signalling of the DNA Damage 3 Response during evolution. 4 5 Yardena Silas 1, 2, Esti Singer 1, Norbert Lehming 2 and Ophry Pines 1, 2* 6 1. Department of Microbiology and Molecular Genetics, IMRIC, Faculty of Medicine, 7 Hebrew University, Jerusalem, Israel 8 2. CREATE‑NUS‑HUJ Program and the Department of Microbiology and Immunology, 9 Yong Loo Lin School of Medicine, National University of Singapore, Singapore, 10 Singapore. 11 12 [email protected] 13 [email protected] 14 [email protected] 15 [email protected] 16 17 18 19 20 21 22 23 24 25 26 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.04.232652; this version posted August 4, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 27 Abstract 28 Class-II fumarase (Fumarate Hydratase, FH) and its metabolic intermediates are essential 29 components in the DNA damage response (DDR) in eukaryotic cells (human and yeast) and 30 in the prokaryote Bacillus subtilis. -

Mutation of the Fumarase Gene in Two Siblings with Progressive Encephalopathy and Fumarase Deficiency T
Mutation of the Fumarase Gene in Two Siblings with Progressive Encephalopathy and Fumarase Deficiency T. Bourgeron,* D. Chretien,* J. Poggi-Bach, S. Doonan,' D. Rabier,* P. Letouze,I A. Munnich,* A. R6tig,* P. Landneu,* and P. Rustin* *Unite de Recherches sur les Handicaps Genetiques de l'Enfant, INSERM U393, Departement de Pediatrie et Departement de Biochimie, H6pital des Enfants-Malades, 149, rue de Sevres, 75743 Paris Cedex 15, France; tDepartement de Pediatrie, Service de Neurologie et Laboratoire de Biochimie, Hopital du Kremlin-Bicetre, France; IFaculty ofScience, University ofEast-London, UK; and IService de Pediatrie, Hopital de Dreux, France Abstract chondrial enzyme (7). Human tissue fumarase is almost We report an inborn error of the tricarboxylic acid cycle, fu- equally distributed between the mitochondria, where the en- marase deficiency, in two siblings born to first cousin parents. zyme catalyzes the reversible hydration of fumarate to malate They presented with progressive encephalopathy, dystonia, as a part ofthe tricarboxylic acid cycle, and the cytosol, where it leucopenia, and neutropenia. Elevation oflactate in the cerebro- is involved in the metabolism of the fumarate released by the spinal fluid and high fumarate excretion in the urine led us to urea cycle. The two isoenzymes have quite homologous struc- investigate the activities of the respiratory chain and of the tures. In rat liver, they differ only by the acetylation of the Krebs cycle, and to finally identify fumarase deficiency in these NH2-terminal amino acid of the cytosolic form (8). In all spe- two children. The deficiency was profound and present in all cies investigated so far, the two isoenzymes have been found to tissues investigated, affecting the cytosolic and the mitochon- be encoded by a single gene (9,10). -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Anti-Inflammatory Role of Curcumin in LPS Treated A549 Cells at Global Proteome Level and on Mycobacterial Infection
Anti-inflammatory Role of Curcumin in LPS Treated A549 cells at Global Proteome level and on Mycobacterial infection. Suchita Singh1,+, Rakesh Arya2,3,+, Rhishikesh R Bargaje1, Mrinal Kumar Das2,4, Subia Akram2, Hossain Md. Faruquee2,5, Rajendra Kumar Behera3, Ranjan Kumar Nanda2,*, Anurag Agrawal1 1Center of Excellence for Translational Research in Asthma and Lung Disease, CSIR- Institute of Genomics and Integrative Biology, New Delhi, 110025, India. 2Translational Health Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, 110067, India. 3School of Life Sciences, Sambalpur University, Jyoti Vihar, Sambalpur, Orissa, 768019, India. 4Department of Respiratory Sciences, #211, Maurice Shock Building, University of Leicester, LE1 9HN 5Department of Biotechnology and Genetic Engineering, Islamic University, Kushtia- 7003, Bangladesh. +Contributed equally for this work. S-1 70 G1 S 60 G2/M 50 40 30 % of cells 20 10 0 CURI LPSI LPSCUR Figure S1: Effect of curcumin and/or LPS treatment on A549 cell viability A549 cells were treated with curcumin (10 µM) and/or LPS or 1 µg/ml for the indicated times and after fixation were stained with propidium iodide and Annexin V-FITC. The DNA contents were determined by flow cytometry to calculate percentage of cells present in each phase of the cell cycle (G1, S and G2/M) using Flowing analysis software. S-2 Figure S2: Total proteins identified in all the three experiments and their distribution betwee curcumin and/or LPS treated conditions. The proteins showing differential expressions (log2 fold change≥2) in these experiments were presented in the venn diagram and certain number of proteins are common in all three experiments. -

Exploring the Non-Canonical Functions of Metabolic Enzymes Peiwei Huangyang1,2 and M
© 2018. Published by The Company of Biologists Ltd | Disease Models & Mechanisms (2018) 11, dmm033365. doi:10.1242/dmm.033365 REVIEW SPECIAL COLLECTION: CANCER METABOLISM Hidden features: exploring the non-canonical functions of metabolic enzymes Peiwei Huangyang1,2 and M. Celeste Simon1,3,* ABSTRACT A key finding from studies of metabolic enzymes is the existence The study of cellular metabolism has been rigorously revisited over the of mechanistic links between their nuclear localization and the past decade, especially in the field of cancer research, revealing new regulation of transcription. By modulating gene expression, insights that expand our understanding of malignancy. Among these metabolic enzymes themselves facilitate adaptation to rapidly insights isthe discovery that various metabolic enzymes have surprising changing environments. Furthermore, they can directly shape a ’ activities outside of their established metabolic roles, including in cell s epigenetic landscape (Kaelin and McKnight, 2013). the regulation of gene expression, DNA damage repair, cell cycle Strikingly, several metabolic enzymes exert completely distinct progression and apoptosis. Many of these newly identified functions are functions in different cellular compartments. Nuclear fructose activated in response to growth factor signaling, nutrient and oxygen bisphosphate aldolase, for example, directly interacts with RNA ́ availability, and external stress. As such, multifaceted enzymes directly polymerase III to control transcription (Ciesla et al., 2014), -

Appendix Table A.2.3.1 Full Table of All Chicken Proteins and Human Orthologs Pool Accession Human Human Protein Human Product Cell Angios Log2( Endo Gene Comp
Appendix table A.2.3.1 Full table of all chicken proteins and human orthologs Pool Accession Human Human Protein Human Product Cell AngioS log2( Endo Gene comp. core FC) Specific CIKL F1NWM6 KDR NP_002244 kinase insert domain receptor (a type III receptor tyrosine M 94 4 kinase) CWT Q8AYD0 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CWT Q8AYD0 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CIKL F1P1Y9 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CIKL F1P1Y9 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CIKL F1N871 FLT4 NP_891555 fms-related tyrosine kinase 4 M 86 -1.71 CWT O73739 EDNRA NP_001948 endothelin receptor type A M 81 -8 CIKL O73739 EDNRA NP_001948 endothelin receptor type A M 81 -8 CWT Q4ADW2 PROCR NP_006395 protein C receptor, endothelial M 80 -0.36 CIKL Q4ADW2 PROCR NP_006395 protein C receptor, endothelial M 80 -0.36 CIKL F1NFQ9 TEK NP_000450 TEK tyrosine kinase, endothelial M 77 7.3 specific CWT Q9DGN6 ECE1 NP_001106819 endothelin converting enzyme 1 M 74 -0.31 CIKL Q9DGN6 ECE1 NP_001106819 endothelin converting enzyme 1 M 74 -0.31 CWT F1NIF0 CA9 NP_001207 carbonic anhydrase IX I 74 CIKL F1NIF0 CA9 NP_001207 carbonic anhydrase IX I 74 CWT E1BZU7 AOC3 NP_003725 amine oxidase, copper containing 3 (vascular adhesion protein M 70 1) CIKL E1BZU7 AOC3 NP_003725 amine oxidase, copper containing 3 (vascular adhesion protein M 70 1) CWT O93419 COL18A1 NP_569712 collagen, type XVIII, alpha 1 E 70 -2.13 CIKL O93419 -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -
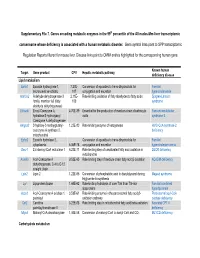
Supplementary File 7. Genes Encoding Metabolic Enzymes in The
Supplementary File 7. Genes encoding metabolic enzymes in the 99th percentile of the All nodes-Mm-liver transcriptomic consensome whose deficiency is associated with a human metabolic disorder. Gene symbol links point to SPP transcriptomic Regulation Reports filtered for mouse liver. Disease links point to OMIM entries highlighted for the corresponding human gene. Known human Target Gene product CPV Hepatic metabolic pathway deficiency disease Lipid metabolism Ephx1 Epoxide hydroxylase 1, 7.27E- Conversion of epoxides to trans-dihydrodiols for Familial microsomal xenobiotic 107 conjugation and excretion hypercholanemia Aldh3a2 Aldehyde dehydrogenase 3 2.11E- Rate-limiting oxidation of fatty aldehydes to fatty acids Sjogren-Larsson family, member A2 (fatty 103 syndrome aldehyde dehydrogenase) Ehhadh Enoyl-Coenzyme A, 4.70E-89 Essential for the production of medium-chain dicarboxylic Fanconi renotubular hydratase/3-hydroxyacyl acids syndrome 3 Coenzyme A dehydrogenase Hmgcs2 3-hydroxy-3-methylglutaryl- 1.23E-80 Rate-limiting enzyme of ketogenesis HMG-CoA synthase-2 coenzyme A synthase 2, deficiency mitochondrial Ephx2 Epoxide hydrolase 2, Conversion of epoxides to trans-dihydrodiols for Familial cytoplasmic 6.06E-78 conjugation and excretion hypercholesterolemia Decr1 2,4-dienoyl-CoA reductase 1 6.23E-71 Rate-limiting step of unsaturated fatty acid oxidation in DECR deficiency mitochondria Acadm Acyl-Coenzyme A 9.52E-65 Rate limiting step of medium-chain fatty acid β-oxidation ACADM deficiency dehydrogenase, C-4 to C-12 straight chain Lpin2