Genetic Portrait of North-West Indian Population Based on X Chromosome Alu Insertion Markers Abstract Alu Insertion Elements
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Any Person May Make a Complaint About The
HIGH COURT OF CHHATTISGARH AT BILASPUR WRIT PETITION (C) No. 4944 of 2009 PETITIONER : Rajendra Singh (Arora). V E R S U S RESPONDENTS : State of Chhattisgarh & Others. PETITION UNDER ARTICLE 226/227 OF THE CONSTITUTION OF INDIA SB: Hon’ble Shri Satish K. Agnihotri, J. --------------------------------------------------------------------------------------------- Present: Shri Kishore Shrivastava, Senior Advocate with Shri Sanjay Tamrakar, Shri Ashish Shirvastava and Shri Anshuman Shrivastava, Advocates for the petitioner. Shri V.V.S.Moorthy, Deputy Advocate General for the State/ respondent No. 1, 2, 4 and 5. Dr. N.K.Shukla, Senior Advocate with Shri Aditya Khare, Advocates for the respondent No. 7 and 8. --------------------------------------------------------------------------------------------- O R D E R (Delivered on 10th day of July, 2013) 1. The petitioner mainly challenges the order dated 23.07.2007 (Annexure P/14) passed by the High Power State Level Caste Scrutiny Committee i.e. respondent No. 3 (for short ‘the Committee’) wherein the petitioner has been found that he does not belong to “Lohar” i.e. Other Backward Class category, and also the subsequent order dated 28.08.2009 (Annexure P/2) wherein on the basis of the order dated 22.09.2008 (Annexure P/1) the petitioner was disqualified for a further period of five years in view of the provisions of section 19(2) of the Chhattisgarh Municipal Corporation Act, 1956 (for short 'the Act, 1956'). 2. The facts, in brief, are that the petitioner, in the year 2000, contested the election as Councillor from Ward No. 22, Municipal 2 Corporation, Bhilai, declaring himself as a member of “Lohar” community that comes within OBC category on the basis of social status certificate dated 10.04.2000 (Annexure P/3). -
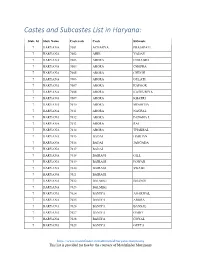
Castes and Subcastes List in Haryana
Castes and Subcastes List in Haryana: State Id State Name Castecode Caste Subcaste 7 HARYANA 7001 ACHARYA PRAJAPATI 7 HARYANA 7002 AHIR YADAV 7 HARYANA 7003 ARORA CHHABRA 7 HARYANA 7004 ARORA CHOPRA 7 HARYANA 7005 ARORA CHUGH 7 HARYANA 7006 ARORA GULATI 7 HARYANA 7007 ARORA KAPOOR 7 HARYANA 7008 ARORA KATHURIYA 7 HARYANA 7009 ARORA KHATRI 7 HARYANA 7010 ARORA MINOCHA 7 HARYANA 7011 ARORA NAGPAL 7 HARYANA 7012 ARORA PANGHA L 7 HARYANA 7013 ARORA RAI 7 HARYANA 7014 ARORA THAKRAL 7 HARYANA 7015 BADAI HARIJAN 7 HARYANA 7016 BADAI JANGADA 7 HARYANA 7017 BADAI 7 HARYANA 7018 BAIRAGI GILL 7 HARYANA 7019 BAIRAGI POWAR 7 HARYANA 7020 BAIRAGI SWAMI 7 HARYANA 7021 BAIRAGI 7 HARYANA 7022 BALMIKI BHANGI 7 HARYANA 7023 BALMIKI 7 HARYANA 7024 BANIYA AGARWAL 7 HARYANA 7025 BANIYA ARORA 7 HARYANA 7026 BANIYA BANSAL 7 HARYANA 7027 BANIYA GARG 7 HARYANA 7028 BANIYA GOYAL 7 HARYANA 7029 BANIYA GUPTA https://www.matchfinder.in/matrimonial/haryana-matrimony This list is provided for free by the courtesy of Matchfinder Matrimony 7 HARYANA 7030 BANIYA JAIN 7 HARYANA 7031 BANIYA JINDAL 7 HARYANA 7032 BANIYA KANSAL 7 HARYANA 7033 BANIYA MAHAJAN 7 HARYANA 7034 BANIYA RANA 7 HARYANA 7035 BANIYA SHAHU 7 HARYANA 7036 BANIYA SINGLA 7 HARYANA 7037 BANIYA 7 HARYANA 7038 BANNSA GARG 7 HARYANA 7039 BAORI 7 HARYANA 7040 BARHAI DHIMAN 7 HARYANA 7041 BARHAI GARG 7 HARYANA 7042 BARHAI KHATI 7 HARYANA 7043 BARHAI SHARMA 7 HARYANA 7044 BARHAI VISHWAKARMA 7 HARYANA 7045 BAWARIA DABLA 7 HARYANA 7046 BAZIGAR BADHAI 7 HARYANA 7047 BHAT ACHARYA 7 HARYANA 7048 BHAT SHARMA 7 HARYANA -

'In Our Whole Society, There Is No Equality': Sikh Householding And
religions Article ‘In Our Whole Society, There Is No Equality’: Sikh Householding and the Intersection of Gender and Caste Nicola Mooney Department of Social, Cultural and Media Studies & South Asian Studies Institute, University of the Fraser Valley, Abbotsford, BC V2S 7M8, Canada; [email protected] Received: 24 December 2019; Accepted: 7 February 2020; Published: 19 February 2020 Abstract: Sikhism is widely understood and celebrated as san egalitarian religion. This follows from its interpretation as a challenge to the caste schema of Hinduism as well as readings which suggest its gender equality. This paper explores the intersection of caste and gender in Sikh society in relation to Guru Nanak’s tenet that Sikhs be householders. Nanak’s view that householding is the basis of religious life and spiritual liberation—as opposed to the caste Hindu framework in which householding relates only to the specific stage of life in which one is married and concerned with domestic affairs—was one of the most important social and ritual reforms he introduced. By eliminating the need for an asceticism supported by householders, or in other words the binary framework of lay and renunciant persons, Nanak envisioned the possibility that the rewards of ascetism could accrue to householders. For Sikhs living at Kartarpur, the first intentional Sikh community, established by Guru Nanak as a place of gathering and meditation, Nanak’s egalitarian ideals were practiced so that women and members of all castes were equal participants. Guru Nanak’s model for social and ritual life presents a radical challenge to the hierarchies and exclusions of Hinduism, and yet, contains within it the basis for ongoing caste and gender disparity for Sikhs, since most Sikhs continue to arrange their householding around caste endogamous marriages and social and domestic arrangements which privilege men. -

Caste, Kinship and Sex Ratios in India
NBER WORKING PAPER SERIES CASTE, KINSHIP AND SEX RATIOS IN INDIA Tanika Chakraborty Sukkoo Kim Working Paper 13828 http://www.nber.org/papers/w13828 NATIONAL BUREAU OF ECONOMIC RESEARCH 1050 Massachusetts Avenue Cambridge, MA 02138 March 2008 We thank Bob Pollak, Karen Norberg, David Rudner and seminar participants at the Work, Family and Public Policy workshop at Washington University for helpful comments and discussions. We also thank Lauren Matsunaga and Michael Scarpati for research assistance and Cassie Adcock and the staff of the South Asia Library at the University of Chicago for their generous assistance in data collection. We are also grateful to the Weidenbaum Center and Washington University (Faculty Research Grant) for research support. The views expressed herein are those of the author(s) and do not necessarily reflect the views of the National Bureau of Economic Research. NBER working papers are circulated for discussion and comment purposes. They have not been peer- reviewed or been subject to the review by the NBER Board of Directors that accompanies official NBER publications. © 2008 by Tanika Chakraborty and Sukkoo Kim. All rights reserved. Short sections of text, not to exceed two paragraphs, may be quoted without explicit permission provided that full credit, including © notice, is given to the source. Caste, Kinship and Sex Ratios in India Tanika Chakraborty and Sukkoo Kim NBER Working Paper No. 13828 March 2008 JEL No. J12,N35,O17 ABSTRACT This paper explores the relationship between kinship institutions and sex ratios in India at the turn of the twentieth century. Since kinship rules varied by caste, language, religion and region, we construct sex-ratios by these categories at the district-level using data from the 1901 Census of India for Punjab (North), Bengal (East) and Madras (South). -

Changing Caste Relations and Emerging Contestations in Punjab
CHANGING CASTE RELATIONS AND EMERGING CONTESTATIONS IN PUNJAB PARAMJIT S. JUDGE When scholars and political leaders characterised Indian society as unity in diversity, there were simultaneous efforts in imagining India as a civilisational unity also. The consequences of this ‘imagination’ are before us in the form of the emergence of religious nationalism that ultimately culminated into the partition of the country. Why have I started my discussion with the issue of religious nationalism and partition? The reason is simple. Once we assume that a society like India could be characterised in terms of one caste hierarchical system, we are essentially constructing the discourse of dominant Hindu civilisational unity. Unlike class and gender hierarchies which are exist on economic and sexual bases respectively, all castes cannot be aggregated and arranged in hierarchy along one axis. Any attempt at doing so would amount to the construction of India as essentially the Hindu India. Added to this issue is the second dimension of hierarchy, which could be seen by separating Varna from caste. Srinivas (1977) points out that Varna is fixed, whereas caste is dynamic. Numerous castes comprise each Varna, the exception to which is the Brahmin caste whose caste differences remain within the caste and are unknown to others. We hardly know how to distinguish among different castes of Brahmins, because there is complete absence of knowledge about various castes among them. On the other hand, there is detailed information available about all the scheduled castes and backward classes. In other words, knowledge about castes and their place in the stratification system is pre- determined by the enumerating agency. -

The Making of the Man's Man: Stardom and the Cultural Politics of Neoliberalism in Hindutva India
Southern Illinois University Carbondale OpenSIUC Dissertations Theses and Dissertations 6-1-2021 THE MAKING OF THE MAN’S MAN: STARDOM AND THE CULTURAL POLITICS OF NEOLIBERALISM IN HINDUTVA INDIA Soumik Pal Southern Illinois University Carbondale, [email protected] Follow this and additional works at: https://opensiuc.lib.siu.edu/dissertations Recommended Citation Pal, Soumik, "THE MAKING OF THE MAN’S MAN: STARDOM AND THE CULTURAL POLITICS OF NEOLIBERALISM IN HINDUTVA INDIA" (2021). Dissertations. 1916. https://opensiuc.lib.siu.edu/dissertations/1916 This Open Access Dissertation is brought to you for free and open access by the Theses and Dissertations at OpenSIUC. It has been accepted for inclusion in Dissertations by an authorized administrator of OpenSIUC. For more information, please contact [email protected]. THE MAKING OF THE MAN’S MAN: STARDOM AND THE CULTURAL POLITICS OF NEOLIBERALISM IN HINDUTVA INDIA by Soumik Pal B.A., Ramakrishna Mission Residential College, Narendrapur, 2005 M.A., Jadavpur University, 2007 PGDM (Communications), Mudra Institute of Communications, Ahmedabad, 2009 A Dissertation Submitted in Partial Fulfillment of the Requirements for the Doctor of Philosophy Degree College of Mass Communication and Media Arts in the Graduate School Southern Illinois University Carbondale May 2021 DISSERTATION APPROVAL THE MAKING OF THE MAN’S MAN: STARDOM AND THE CULTURAL POLITICS OF NEOLIBERALISM IN HINDUTVA INDIA by Soumik Pal A Dissertation Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in the field of Mass Communication and Media Arts Approved by: Dr. Jyotsna Kapur, Chair Dr. Walter Metz Dr. Deborah Tudor Dr. Novotny Lawrence Dr. -

"Denotified" Tribes (Vimukt Jatis) of Punjab (North-West India) and Its Social Implication
Final Report on UGC Major Research Project (MRP) Anthropogenetic Profile of "denotified" Tribes (Vimukt Jatis) of Punjab (North-West India) and its Social Implication (From 01/ 04/ 2013 to 31/ 03/ 2017) Submitted to The University Grants Commission, New Delhi by Prof. (Dr.) S. M .S. Chahal Principal Investigator Department of Human Genetics (earlier Department of Human Biology) Punjabi University, Patiala 147002 (Punjab) "Anthropogenetic Profile of "denotified" Tribes (Vimukt Jatis) of Punjab (North-West India) and its Social Implication" INTRODUCTION The “denotified” tribes, also known as the Vimukt Jatis, are the tribes that were originally listed under the Criminal Tribes Act of 1871, as Criminal Tribes and "addicted to the systematic commission of non-bailable offences." Once a tribe became "notified" as criminal, all its members were required to register with the local magistrate, failing which they would be charged with a crime under the Indian Penal Code. The Act was repealed in August 1949 and former "criminal tribes" were “denotified” in 1952, when the Act was replaced with the Habitual Offenders Act 1952 of Government of India, and in 1961 state governments started releasing lists of such tribes. At the time of the independence in 1947, there were 13 million people in 127 communities who faced constant surveillance, search and arrest without warrant if any member of the group was found outside the prescribed area. The creation of these categories should be seen in the context of colonialism. The British authorities listed them separately by creating a category of castes or tribes labeled as criminal. The first Census of India was conducted in 1871 and at that time there was no consensus or any definition of "tribe". -

Government of Punjab
GOVERNMENT OF PUNJAB TELEPHONE DIRECTORY-2016 PUBLIC WORKS DEPARTMENT (B&R) BR. CONTENTS Page No. PUNJAB RAJ BHAWAN 1 PUNJAB VIDHAN SABHA 1-2 PUNJAB & HARYANA HIGH COURT 2-5 CHIEF MINISTER OFFICE 5-8 DEPUTY CHIEF MINISTER OFFICE 8-9 CABINET MINISTERS 9-12 CHIEF PARLIAMENTARY SECRETARIES 12-14 LEADER OF OPPOSITION 14 MEMBERS OF PARLIAMENT FROM PUNJAB 15-17 MEMBERS OF PUNJAB LEGISLATIVE ASSEMBLY 17-24 POLITICAL PARTIES IN PUNJAB VIDHAN SABHA 25 LOK PAL 25 CHIEF SECRETARY 25 ADVOCATE GENERAL 25 COMMISSIONS 26-32 FINANCIAL COMMISSIONERS/PRINCIPAL SECRETARIES 32-34 ADMINISTRATIVE SECRETARIES 34-36 SPECIAL SECRETARIES 36-37 ADDITIONAL SECRETARIES 37-38 JOINT SECRETARIES 38-39 HEAD OF DEPARTMENTS 39-48 INFORMATION AND PUBLIC RELATIONS 48-51 POLICE DEPARTMENT 51-54 CORPORATIONS / BOARDS 55-68 COMMISSIONERS OF DIVISIONS 68 DEPUTY COMMISSIONERS 68-69 MAYORS & COMMISSIONERS OF MUNICIPAL CORPORATIONS 69-70 CHAIRMAN DISTT. PLANNING COMMITTEE 71 UNIVERSITIES 72 PRESIDENT SECRETARIAT 73 VICE PRESIDENT SECRETARIAT 73 SUPREME COURT OF INDIA 73 ELECTION COMMISSION OF INDIA 73 NITI AAYOG 73 PRIME MINISTER OF INDIA 74 PARLIAMENT (RAJYA SABHA) 74 PARLIAMENT (LOK SABHA) 74 IAS & OTHER OFFICERS OF PB. POSTED AT DELHI 75 RESIDENT COMMISSIONER OFFICE AT NEW DELHI 76-77 CHANDIGARH ADMINISTRATION 77-78 OTHERS 78-80 PRESS, RADIO & TV 80-120 GUEST HOUSE/ENTERTAINMENT CENTRE (CHD) 120-121 CIRCUIT HOUSES IN PUNJAB & H.P. 121 EMERGENCY & GEN. UTILITY TEL. NO. AT CHD 121-123 SOME IMPORTANT TELEPHONE NUMBERS AT DELHI 123-124 WEBSITES OF VARIOUS PUNJAB GOVT. DEPARTMENTS BOARDS, COPORATIONS, ETC. 124-125 OFFICIAL EMAIL ID’S OF THE DEPARTMENTS 125-132 STD CODES OF CITIES INDIA 138-143 *OEFY 1 Name & Designation Phone Residence Off. -

Militarization of Sikh Masculinity Aakriti Kohli
Militarization of Sikh Masculinity Aakriti Kohli ABSTRACT: Critically reading the theoretical and descriptive scholarly work on colonial Punjab, Sikhs, Sikhism and the imperial British Empire, this paper traces how the formation of Sikh martial masculinity rooted in religious tradition was institutionalized into a particular form of militarized masculinity in the colonial period in Punjab, India. Additionally, it explores how the historical construction of masculinity intersects with the contemporary discourses on Sikh identity and masculinity in the diaspora, specifically in Britain. With reference to British Sikhs and their project of reclaiming recognition of their contribution in WWI, the paper goes on to argue that perhaps the projection of Khalsa identity as synonymous with Sikh identity and the performance of Sikh masculinity lies in projecting and representing themselves as warriors, to seek legitimacy from the military of their masculinity in exhibiting war effort. KEYWORDS: masculinity, military, martial, Sikhs, Punjab, Khalsa The dominant perception of Sikhs as martial, brave and willing to sacrifice is re- flected in popular culture at large. By extension and association, Punjab, seen as the homeland of Sikhs, finds itself venerated as the land of the brave, or the land of the lions, if you like. This idea of the Sikh identity and Sikh masculinity in particular is a very real form of consciousness which defines, shapes and configures Sikh masculinity and performance of the male self, and are ideas in which many Sikh men root their identity. As I have argued elsewhere, this particular masculine per- formance does draw its strength from religious rituals and practices.1 It might not Graduate Journal of Social Science November 2016, Vol. -

Dakhani Sikhs and the Legacy of Guru Gobind Singh
31 Singh: Dakhani Sikhs Dakhani Sikhs and the Legacy of Guru Gobind Singh Birinder Pal Singh Punjabi University, Patiala, Punjab _______________________________________________________________ The Dakhani Sikhs are residents of the Nizam of Hyderabad’s state of Deccan now trifurcated into three states of Telengana, Maharashtra and Karnataka. Hyderabad and Nanded are the cities with largest population concentration of Sikhs. They claim to be the descendants of the Lahori Fauj of Maharaja Ranjit Singh that arrived in Hyderabad in 1830 at the Nizam’s request to restore law and order in the state. They married local women and adopted local cultural and religious practices too without compromising Sikh tenets. They are particular about their Sikh form and identity and claim to carry the legacy of Guru Gobind Singh who spent the last days of his life at Nanded. They believe themselves to be sustaining the Sikh religion, though the Punjabi Sikhs call them ‘duplicate’ Sikhs. _______________________________________________________________ Two centuries before the last guru, Baba Nanak traversed the terrains of south India including Bidar where now stands Gurdwara Nanak Jhira, an important centre in the Sikh religious itinerary of Nanded. The latter place, however, is known for the last guru’s arrival in the Deccan, and departure to the other world. The overarching pre-eminence of Sach Khand Sri Hazoor Sahib Abchalnagar (gurdwara’s name) at Nanded is more than conspicuous, and so is the influence and significance of Guru Gobind Singh associated with it. It is one of the five seats of Sikh religious authority (takht) and the only one in south India. It assumes importance for the Sikhs since the tenth and the last living guru not only spent the last days of his life there but bestowed gurudom to the Sikh scripture, the Granth Sahib, ordaining: Sab Sikhan ko huqam hai, guru maniyo Granth. -

The Char Bagh-I Panjab: Socio-Cultural Configuration
23 J. S. Grewal: Char-i Bagh Panjab The Char Bagh-i Panjab: Socio-Cultural Configuration J.S. Grewal Institute of Punjab Studies, Chandigarh _______________________________________________________________ The descriptive part of the mid-nineteenth century Char Bagh-i Punjab of Ganesh Das in Persian is analysed for evidence on the social and cultural history of the Punjab during the eighteenth and the early nineteenth century. The disparate facts and places in specific space and time by the author are sought to be classified and put together coherently in terms of urbanization; religious beliefs, practices, and institutions of Hindus, Muslims and Sikhs; popular religion; traditional learning, both religious and secular; literature in Persian, Punjabi, Braj and ‘Hindi’ in Perso-Arabic, Gurmukhi and Devanagri scripts; and gender relations, with special regard to conjugal and personal love. The socio-cultural configuration that emerges on the whole from this analysis is given at the end, with the reflection of the author’s personality as a member of the precolonial Punjabi society. Important in itself, this configuration provides the background for an appreciation of socio-cultural change in the colonial Punjab. _______________________________________________________________ Introducton The Char Bagh-i Punjab of Ganesh Das Badhera is by far the most important single source of information for the social and cultural history of the Punjab before its annexation to British India in 1849.* Though a general history from the earliest times to the fall of the kingdom of Lahore in 1849, about three- fourths of the work is devoted to the eighteenth and the early nineteenth century, both in its narrative part which is almost entirely political, and its descriptive part which is almost entirely non-political. -

Estimated Population by Castes, 21 Punjab
ES.TIMATED POP'ULATION BY CASTES .. 1951 21. PUNJAB Office 0/ the Registrar General, India MINISTRY OF HOME AFFAIRS GOVERNlv1ENT OF INDIA I 954 INTRODUC'I'ION._--- In pursuance of Government policy there WaS limited enumerAtion and tabulation of Qastes in 1951 Census. Bven in the case of Scheduled Castes, Scheduled Tribes andoackM Ward Classe~ the figures of each caste were not separately extracted; only the group totals were ascertained. The "Backward Classes Commission require the figures of population of each individual caste. In order to assist them an estimate - of population of each caste in IS51 has been made on the basis of the figures of the previous censuses •. 2. The figures have been presented in three taDles:- (i) Scheduled Castes, Hindus only (i1) Scheduled Tribes -(iii) Other Castes, Hindus and Muslims separately. 3. No castewise figures are available for 1841 Census. The tables of 1£41 Census giye figures for. only a rew/castes and these also fo~ a few seleeted districts. 4. Extracts frGm previous censuses Reports of undivided Punjab, explaining the causes for variation in the figures of individual caste have been given in an Appendix, TABLE I - SCHEDULED CASTES The figures given in this table relate to the territory of Punjab as in 1951. 2. The table presents the figures of 34 castes as specified in the Presidentts Order of 1950. The population of each caste given in this table refers only to the popula tion of Hindus. 3. Column 5 of the table gives the estimated popUlation in ISSI. This has been determined by applying the percentage increase of the general pop~lation of the state to the latest available census figures of each caste.