Neural Stem Cell-Derived Exosomes Revert HFD-Dependent Memory Impairment Via CREB-BDNF Signalling
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chr21 Protein-Protein Interactions: Enrichment in Products Involved in Intellectual Disabilities, Autism and Late Onset Alzheimer Disease
bioRxiv preprint doi: https://doi.org/10.1101/2019.12.11.872606; this version posted December 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Chr21 protein-protein interactions: enrichment in products involved in intellectual disabilities, autism and Late Onset Alzheimer Disease Julia Viard1,2*, Yann Loe-Mie1*, Rachel Daudin1, Malik Khelfaoui1, Christine Plancon2, Anne Boland2, Francisco Tejedor3, Richard L. Huganir4, Eunjoon Kim5, Makoto Kinoshita6, Guofa Liu7, Volker Haucke8, Thomas Moncion9, Eugene Yu10, Valérie Hindie9, Henri Bléhaut11, Clotilde Mircher12, Yann Herault13,14,15,16,17, Jean-François Deleuze2, Jean- Christophe Rain9, Michel Simonneau1, 18, 19, 20** and Aude-Marie Lepagnol- Bestel1** 1 Centre Psychiatrie & Neurosciences, INSERM U894, 75014 Paris, France 2 Laboratoire de génomique fonctionnelle, CNG, CEA, Evry 3 Instituto de Neurociencias CSIC-UMH, Universidad Miguel Hernandez-Campus de San Juan 03550 San Juan (Alicante), Spain 4 Department of Neuroscience, The Johns Hopkins University School of Medicine, Baltimore, MD 21205 USA 5 Center for Synaptic Brain Dysfunctions, Institute for Basic Science, Daejeon 34141, Republic of Korea 6 Department of Molecular Biology, Division of Biological Science, Nagoya University Graduate School of Science, Furo, Chikusa, Nagoya, Japan 7 Department of Biological Sciences, University of Toledo, Toledo, OH, 43606, USA 8 Leibniz Forschungsinstitut für Molekulare Pharmakologie -

The B Vitamins Nicotinamide (B3) and Riboflavin (B2)
The B Vitamins Nicotinamide (B3) and Riboflavin (B2) Stimulate Metamorphosis in Larvae of the Deposit- Feeding Polychaete Capitella teleta: Implications for a Sensory Ligand-Gated Ion Channel Robert T. Burns1*, Jan A. Pechenik1, William J. Biggers2, Gia Scavo2, Christopher Lehman2 1 Department of Biology, Tufts University, Medford, Massachusetts, United States of America, 2 Department of Biology, Wilkes University, Wilkes-Barre, Pennsylvania, United States of America Abstract Marine sediments can contain B vitamins, presumably incorporated from settled, decaying phytoplankton and microorganisms associated with decomposition. Because B vitamins may be advantageous for the energetically intensive processes of metamorphosis, post-metamorphic growth, and reproduction, we tested several B vitamins to determine if they would stimulate larvae of the deposit-feeding polychaete Capitella teleta to settle and metamorphose. Nicotinamide and riboflavin individually stimulated larvae of C. teleta to settle and metamorphose, generally within 1–2 hours at nicotinamide concentrations as low as 3 mM and riboflavin concentrations as low as 50 mM. More than 80% of the larvae metamorphosed within 30 minutes at a nicotinamide concentration of 7 mM. The pyridine channel agonist pyrazinecarboxamide also stimulated metamorphosis at very low concentrations. In contrast, neither lumichrome, thiamine HCl, pyridoxine HCl, nor vitamin B12 stimulated larvae of C. teleta to metamorphose at concentrations as high as 500 mM. Larvae also did not metamorphose in response to either nicotinamide or pyrazinecarboxamide in calcium-free seawater or with the addition of 4-acetylpyridine, a competitive inhibitor of the pyridine receptor. Together, these results suggest that larvae of C. teleta are responding to nicotinamide and riboflavin via a chemosensory pyridine receptor similar to that previously reported to be present on crayfish chela and involved with food recognition. -

Neurotransmitters-Drugs Andbrain Function.Pdf
Neurotransmitters, Drugs and Brain Function. Edited by Roy Webster Copyright & 2001 John Wiley & Sons Ltd ISBN: Hardback 0-471-97819-1 Paperback 0-471-98586-4 Electronic 0-470-84657-7 Neurotransmitters, Drugs and Brain Function Neurotransmitters, Drugs and Brain Function. Edited by Roy Webster Copyright & 2001 John Wiley & Sons Ltd ISBN: Hardback 0-471-97819-1 Paperback 0-471-98586-4 Electronic 0-470-84657-7 Neurotransmitters, Drugs and Brain Function Edited by R. A. Webster Department of Pharmacology, University College London, UK JOHN WILEY & SONS, LTD Chichester Á New York Á Weinheim Á Brisbane Á Singapore Á Toronto Neurotransmitters, Drugs and Brain Function. Edited by Roy Webster Copyright & 2001 John Wiley & Sons Ltd ISBN: Hardback 0-471-97819-1 Paperback 0-471-98586-4 Electronic 0-470-84657-7 Copyright # 2001 by John Wiley & Sons Ltd. Bans Lane, Chichester, West Sussex PO19 1UD, UK National 01243 779777 International ++44) 1243 779777 e-mail +for orders and customer service enquiries): [email protected] Visit our Home Page on: http://www.wiley.co.uk or http://www.wiley.com All Rights Reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording, scanning or otherwise, except under the terms of the Copyright, Designs and Patents Act 1988 or under the terms of a licence issued by the Copyright Licensing Agency Ltd, 90 Tottenham Court Road, London W1P0LP,UK, without the permission in writing of the publisher. Other Wiley Editorial Oces John Wiley & Sons, Inc., 605 Third Avenue, New York, NY 10158-0012, USA WILEY-VCH Verlag GmbH, Pappelallee 3, D-69469 Weinheim, Germany John Wiley & Sons Australia, Ltd. -

Investigation of Candidate Genes and Mechanisms Underlying Obesity
Prashanth et al. BMC Endocrine Disorders (2021) 21:80 https://doi.org/10.1186/s12902-021-00718-5 RESEARCH ARTICLE Open Access Investigation of candidate genes and mechanisms underlying obesity associated type 2 diabetes mellitus using bioinformatics analysis and screening of small drug molecules G. Prashanth1 , Basavaraj Vastrad2 , Anandkumar Tengli3 , Chanabasayya Vastrad4* and Iranna Kotturshetti5 Abstract Background: Obesity associated type 2 diabetes mellitus is a metabolic disorder ; however, the etiology of obesity associated type 2 diabetes mellitus remains largely unknown. There is an urgent need to further broaden the understanding of the molecular mechanism associated in obesity associated type 2 diabetes mellitus. Methods: To screen the differentially expressed genes (DEGs) that might play essential roles in obesity associated type 2 diabetes mellitus, the publicly available expression profiling by high throughput sequencing data (GSE143319) was downloaded and screened for DEGs. Then, Gene Ontology (GO) and REACTOME pathway enrichment analysis were performed. The protein - protein interaction network, miRNA - target genes regulatory network and TF-target gene regulatory network were constructed and analyzed for identification of hub and target genes. The hub genes were validated by receiver operating characteristic (ROC) curve analysis and RT- PCR analysis. Finally, a molecular docking study was performed on over expressed proteins to predict the target small drug molecules. Results: A total of 820 DEGs were identified between -

Understanding Allergic Multimorbidity Within the Non-Eosinophilic
University of Groningen Understanding allergic multimorbidity within the non-eosinophilic interactome Aguilar, Daniel; Lemonnier, Nathanael; Koppelman, Gerard H; Melén, Erik; Oliva, Baldo; Pinart, Mariona; Guerra, Stefano; Bousquet, Jean; Anto, Josep M Published in: PLoS ONE DOI: 10.1371/journal.pone.0224448 IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document version below. Document Version Publisher's PDF, also known as Version of record Publication date: 2019 Link to publication in University of Groningen/UMCG research database Citation for published version (APA): Aguilar, D., Lemonnier, N., Koppelman, G. H., Melén, E., Oliva, B., Pinart, M., Guerra, S., Bousquet, J., & Anto, J. M. (2019). Understanding allergic multimorbidity within the non-eosinophilic interactome. PLoS ONE, 14(11), [e0224448]. https://doi.org/10.1371/journal.pone.0224448 Copyright Other than for strictly personal use, it is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license (like Creative Commons). Take-down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Downloaded from the University of Groningen/UMCG research database (Pure): http://www.rug.nl/research/portal. For technical reasons the number of authors shown on this cover page is limited to 10 maximum. Download date: 26-12-2020 RESEARCH ARTICLE Understanding allergic multimorbidity within the non-eosinophilic interactome 1,2,3 4 5,6 7 Daniel AguilarID *, Nathanael LemonnierID , Gerard H. -

BDNF Integrity in Ageing and Stress
MOJ Gerontology & Geriatrics Editorial Open Access BDNF integrity in ageing and stress Editorial Volume 1 Issue 6 - 2017 Alzheimer’s disease (AD) presents a progressive, stage-dependent Trevor Archer age-related neurodegenerative disorder that is characterized by Department of Psychology, University of Gothenburg, Sweden aggregation of toxic forms of amyloid β peptide (Aβ). There is much support for the notion that that brain-derived neurotrophic factor Correspondence: Trevor Archer, Department of Psychology, (BDNF) mediates beneficial effects of exercise on neuroplasticity and University of Gothenburg, Sweden, cellular stress resistance.1‒3 BDNF is associated with neuroplasticity Email [email protected] changes promoting health and well-being whereas these influences Received: July 16, 2017 | Published: August 01, 2017 may be opposed by pro-inflammatory cytokines, key factors in neurodegenerative processes.4 Lower serum levels are linked to greater symptom, cognitive, impairment.5 Thus, interventions, as for example physical exercise, whether acute or chronic, endurance or resistance, promote the mobilization of BDNF and other neurotrophic factors6,7 increase hippocampal and other brain regional resilience8 and therewith progression under conditions of stress when confronted critical life protect against cell death by inducing cell proliferation and maturation episodes or periods, appears to be modulated by the availability and with enhanced neuronal reparation, neurogenesis and the growth and accessibility of BDNF.20‒22 Over a wide range of neurologic and functioning of neurons in neurodegenerative disorders.9,10 Sampaio et psychiatric disease states it is found that the integrity and concentrations al.11 in a review of the therapeutic applications of neurotrophic factors, of brain, serum and plasma BDNF is compromised, particularly under particularly BDNF, postulate that they are essential for survival, stressful conditions. -
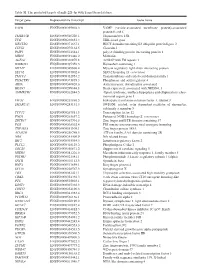
Table SI. the Predicted Targets of Mir-221-3P with Targetscan Database
Table SI. The predicted targets of miR-221-3p with TargetScan database. Target gene Representative transcript Gene name VAPB ENST00000395802.3 VAMP (vesicle-associated membrane protein)-associated protein B and C TMSB15B ENST00000540220.1 Thymosin beta 15B TFG ENST00000240851.4 TRK-fused gene HECTD2 ENST00000371667.1 HECT domain containing E3 ubiquitin protein ligase 2 CLVS2 ENST00000275162.5 Clavesin 2 PAIP1 ENST00000514514.1 poly(A) binding protein interacting protein 1 MIDN ENST00000591446.2 Midnolin AGFG1 ENST00000310078.8 ArfGAP with FG repeats 1 HMBOX1 ENST00000397358.3 Homeobox containing 1 MYLIP ENST00000349606.4 Myosin regulatory light chain interacting protein SEC62 ENST00000337002.4 SEC62 homolog (S. cerevisiae) TMCC1 ENST00000432054.2 Transmembrane and coiled-coil domain family 1 PHACTR4 ENST00000373839.3 Phosphatase and actin regulator 4 AIDA ENST00000340020.6 Axin interactor, dorsalization associated BEAN1 ENST00000299694.8 Brain expressed, associated with NEDD4, 1 AMMECR1 ENST00000262844.5 Alport syndrome, midface hypoplasia and elliptocytosis chro- mosomal region gene 1 EIF3J ENST00000261868.5 Eukaryotic translation initiation factor 3, subunit J SMARCA5 ENST00000283131.3 SWI/SNF related, actin dependent regulator of chromatin, subfamily a, member 5 TCF12 ENST00000267811.5 Transcription factor 12 PNO1 ENST00000263657.2 Partner of NOB1 homolog (S. cerevisiae) ZBTB37 ENST00000367701.5 Zinc finger and BTB domain containing 37 FOS ENST00000303562.4 FBJ murine osteosarcoma viral oncogene homolog ZNF385A ENST00000551109.1 Zinc -

This Thesis Has Been Submitted in Fulfilment of the Requirements for a Postgraduate Degree (E.G
This thesis has been submitted in fulfilment of the requirements for a postgraduate degree (e.g. PhD, MPhil, DClinPsychol) at the University of Edinburgh. Please note the following terms and conditions of use: This work is protected by copyright and other intellectual property rights, which are retained by the thesis author, unless otherwise stated. A copy can be downloaded for personal non-commercial research or study, without prior permission or charge. This thesis cannot be reproduced or quoted extensively from without first obtaining permission in writing from the author. The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the author. When referring to this work, full bibliographic details including the author, title, awarding institution and date of the thesis must be given. The CX3CR1/CX3CL1 Axis Drives the Migration and Maturation of Oligodendroglia in the Central Nervous System Catriona Ford Thesis Submitted for the Degree of Doctor of Philosophy The University of Edinburgh 2017 Abstract In the central nervous system, the axons of neurons are protected from damage and aided in electrical conductivity by the myelin sheath, a complex of proteins and lipids formed by oligodendrocytes. Loss or damage to the myelin sheath may result in impairment of electrical axonal conduction and eventually to neuronal death. Such demyelination is responsible, at least in part, for the disabling neurodegeneration observed in pathologies such as Multiple Sclerosis (MS) and Spinal Cord Injury. In the regenerative process of remyelination, oligodendrocyte precursor cells (OPCs), the resident glial stem cell population of the adult CNS, migrate toward the injury site, proliferate and differentiate into adult oligodendrocytes which subsequently reform the myelin sheath. -

Supplemental Figures
Supplemental Figures Supplemental figure legends Figure S1 | Testing the pre-clustering heuristic. (A) (Left) Default, unsupervised heuristic sets a cut of 7% of the total dendrogram depth, which results in 52 pre-clusters. (Right) The numerical model calculated using the 52 pre-clusters. Xc1 and Xc2 represent the expression (in a binned UMIs grid) of a given gene X in two cells c1 and c2 belonging to the same pre-cluster. The cumulative distribution plot estimates the frequency, hence likelihood, of an expression change. (B) (Left) Forcing a cut of only 4% creates 1152 pre-clusters, more than 20-fold increase compared to the default 7% depth. Also, given the reduction of the average cluster size and the consequent reduction of possible intra-cluster pair-wise comparison, the number of data points used to fit the model decreases of more than 5-fold compared to default 7% cut (from 3.79E+9 to 6.56E+8). (Right) Despite this, the difference between the numerical model of 4% cut and 7% cut is marginal. (C) (Left) Forcing a cut of 20% creates only 9 pre-clusters, which is less than the number of final clusters (in this case, 11) and therefore represents a miscalculated configuration. Still the difference between the numerical model of 20% cut and 7% cut is marginal (right). (D) Also switching from Pearson to Spearman correlation is associated with neglectable differences in the numerical model. (E) (Top) Number of pre-clusters associated with the different cutting depths, correlations metrics (Pearson, Spearman) or linkage metrics (complete or Weighted average distance, WPGMA, instead of default Ward’s). -

Supplementary Table 8. Cpcp PPI Network Details for Significantly Changed Proteins, As Identified in 3.2, Underlying Each of the Five Functional Domains
Supplementary Table 8. cPCP PPI network details for significantly changed proteins, as identified in 3.2, underlying each of the five functional domains. The network nodes represent each significant protein, followed by the list of interactors. Note that identifiers were converted to gene names to facilitate PPI database queries. Functional Domain Node Interactors Development and Park7 Rack1 differentiation Kcnma1 Atp6v1a Ywhae Ywhaz Pgls Hsd3b7 Development and Prdx6 Ncoa3 differentiation Pla2g4a Sufu Ncf2 Gstp1 Grin2b Ywhae Pgls Hsd3b7 Development and Atp1a2 Kcnma1 differentiation Vamp2 Development and Cntn1 Prnp differentiation Ywhaz Clstn1 Dlg4 App Ywhae Ywhab Development and Rac1 Pak1 differentiation Cdc42 Rhoa Dlg4 Ctnnb1 Mapk9 Mapk8 Pik3cb Sod1 Rrad Epb41l2 Nono Ltbp1 Evi5 Rbm39 Aplp2 Smurf2 Grin1 Grin2b Xiap Chn2 Cav1 Cybb Pgls Ywhae Development and Hbb-b1 Atp5b differentiation Hba Kcnma1 Got1 Aldoa Ywhaz Pgls Hsd3b4 Hsd3b7 Ywhae Development and Myh6 Mybpc3 differentiation Prkce Ywhae Development and Amph Capn2 differentiation Ap2a2 Dnm1 Dnm3 Dnm2 Atp6v1a Ywhab Development and Dnm3 Bin1 differentiation Amph Pacsin1 Grb2 Ywhae Bsn Development and Eef2 Ywhaz differentiation Rpgrip1l Atp6v1a Nphp1 Iqcb1 Ezh2 Ywhae Ywhab Pgls Hsd3b7 Hsd3b4 Development and Gnai1 Dlg4 differentiation Development and Gnao1 Dlg4 differentiation Vamp2 App Ywhae Ywhab Development and Psmd3 Rpgrip1l differentiation Psmd4 Hmga2 Development and Thy1 Syp differentiation Atp6v1a App Ywhae Ywhaz Ywhab Hsd3b7 Hsd3b4 Development and Tubb2a Ywhaz differentiation Nphp4 -

Identification of Key Pathways and Genes in Dementia Via Integrated Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.18.440371; this version posted July 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Identification of Key Pathways and Genes in Dementia via Integrated Bioinformatics Analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karnataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2021.04.18.440371; this version posted July 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract To provide a better understanding of dementia at the molecular level, this study aimed to identify the genes and key pathways associated with dementia by using integrated bioinformatics analysis. Based on the expression profiling by high throughput sequencing dataset GSE153960 derived from the Gene Expression Omnibus (GEO), the differentially expressed genes (DEGs) between patients with dementia and healthy controls were identified. With DEGs, we performed a series of functional enrichment analyses. Then, a protein–protein interaction (PPI) network, modules, miRNA-hub gene regulatory network and TF-hub gene regulatory network was constructed, analyzed and visualized, with which the hub genes miRNAs and TFs nodes were screened out. Finally, validation of hub genes was performed by using receiver operating characteristic curve (ROC) analysis. -

The Metabotropic Receptor Mglur6 May Signal Through Go, but Not Phosphodiesterase, in Retinal Bipolar Cells
The Journal of Neuroscience, April 15, 1999, 19(8):2938–2944 The Metabotropic Receptor mGluR6 May Signal Through Go, But Not Phosphodiesterase, in Retinal Bipolar Cells Scott Nawy Departments of Ophthalmology and Visual Science, and Neuroscience, Albert Einstein College of Medicine, Bronx, New York 10461 Bipolar cells are retinal interneurons that receive synaptic input with cells dialyzed with cGMP alone. Comparable results were from photoreceptors. Glutamate, the photoreceptor transmitter, obtained with the PDE inhibitor 3-isobutyl-1-methyl-xanthine hyperpolarizes On bipolar cells by closing nonselective cation (IBMX) or with 8-pCPT-cGMP and IBMX together, indicating channels, an effect mediated by the metabotropic receptor that PDE is not required for mGluR6 signal transduction. Addi- a mGluR6. Previous studies of mGluR6 transduction have sug- tion of the G-protein subunit Go to the pipette solution sup- gested that the receptor couples to a phosphodiesterase (PDE) pressed the cation current and occluded the glutamate re- a bg that preferentially hydrolyzes cGMP, and that cGMP directly gates sponse, whereas dialysis with Gi or with transducin G had the nonselective cation channel. This hypothesis was tested by no significant effect on either the cation current or the response. a dialyzing On bipolar cells with nonhydrolyzable analogs of cGMP. Dialysis of an antibody directed against Go also reduced the Whole-cell recordings were obtained from On bipolar cells in slices glutamate response, indicating a functional role for endoge- a of larval tiger salamander retina. Surprisingly, On bipolar cells nous Go . These results indicate that mGluR6 may signal dialyzed with 8-(4-chlorophenylthio)-cyclic GMP (8-pCPT-cGMP), through Go , rather than a transducin-like G-protein.