International Application Number: Published: PCT/US20 19/052026 — with International Search Report (Art
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Non-Canonical Activation of CREB Mediates Neuroprotection in a C
bioRxiv preprint doi: https://doi.org/10.1101/261420; this version posted February 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Non-Canonical Activation of CREB Mediates Neuroprotection in a C. elegans Model of Excitotoxic Necrosis K. Genevieve Feldmann1,2, Ayesha Chowdhury1,2, Jessi Becker3, N’Gina McAlpin3, Taqwa Ahmed4, Syed Haider3, Jian X. Richard Xia4, Karina Diaz4, Monal G. Mehta5, and Itzhak Mano1,2,4,CA 1 Department of Molecular, Cellular and Biomedical Sciences, CDI Cluster on Neural Development and Repair, The CUNY School of Medicine, City College (CCNY), The City University of New York (CUNY) 2 The CUNY Neuroscience Collaborative PhD Program, CUNY Graduate Center 3 Undergraduate Program in Biology, CCNY, CUNY 4 The Sophie Davis BS/MD program, CUNY School of Medicine 5 Robert Wood Johnson Medical School, Rutgers – The State University of New Jersey CA Corresponding Author: Itzhak Mano, Ph.D. Department of Molecular, Cellular and Biomedical Sciences Center for Discovery & Innovation, Cluster on Neural Development and Repair The CUNY School of Medicine at City College & The CUNY Graduate Center The City University of New York CDI building room 3-382 85 St. Nicholas Terrace, New York, NY 10031 E-mail: [email protected] Office Phone:(212) 6507965 Lab Phone:(212) 6505334 www.manolab.org Running Title: Non-canonical CREB activation in nematode excitotoxicity 1 bioRxiv preprint doi: https://doi.org/10.1101/261420; this version posted February 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

The Coactivator CRTC1 Promotes Cell Proliferation and Transformation Via AP-1
The coactivator CRTC1 promotes cell proliferation and transformation via AP-1 Gianluca Canettieria,1, Sonia Conia,2, Michele Della Guardiaa,2, Valentina Nocerinoa, Laura Antonuccia, Laura Di Magnoa, Robert Screatonc, Isabella Screpantia,b, Giuseppe Gianninia, and Alberto Gulinoa,b,d,1 aDepartment of Experimental Medicine and bPasteur Institute, Cenci Bolognetti Foundation, Sapienza University, Rome, Italy; cApoptosis Research Centre, Children’s Hospital of Eastern Ontario, Ottawa, ON, Canada; and dNeuromed Institute, Pozzilli, Italy Edited by Peter K. Vogt, Scripps Research Institute, La Jolla, CA, and approved December 5, 2008 (received for review September 3, 2008) Regulation of gene expression in response to mitogenic stimuli is a Besides TPA, AP-1 also is activated by growth factors, cytokines, critical aspect underlying many forms of human cancers. The AP-1 stress signals, and oncoproteins, and thus is involved in various complex mediates the transcriptional response to mitogens, and its cellular processes, including cell proliferation, differentiation, deregulation causes developmental defects and tumors. We report cell survival, apoptosis, and neoplastic transformation (14). that the coactivator CRTC1 cyclic AMP response element-binding Whereas the biological role of the AP-1 transcription factors protein (CREB)-regulated transcription coactivator 1 is a potent and is well established, the molecular mechanism through which indispensable modulator of AP-1 function. After exposure of cells to these factors promote target gene activation remains incom- the AP-1 agonist 12-O-tetradecanoylphorbol-13-acetate (TPA), CRTC1 pletely understood. In this report, we demonstrate that CRTC1 is recruited to AP-1 target gene promoters and associates with c-Jun is a potent, indispensable modulator of AP-1 activity that and c-Fos to activate transcription. -

Role of CREB/CRTC1-Regulated Gene Transcription During Hippocampal-Dependent Memory in Alzheimer’S Disease Mouse Models
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Institut de Neurociències Universitat Autònoma de Barcelona Departament de Bioquímica i Biologia Molecular Unitat de Bioquímica, Facultat de Medicina Role of CREB/CRTC1-regulated gene transcription during hippocampal-dependent memory in Alzheimer’s disease mouse models Arnaldo J. Parra Damas TESIS DOCTORAL Bellaterra, 2015 Institut de Neurociències Departament de Bioquímica i Biologia Molecular Universitat Autònoma de Barcelona Role of CREB/CRTC1-regulated gene transcription during hippocampal-dependent memory in Alzheimer’s disease mouse models Papel de la transcripción génica regulada por CRTC1/CREB durante memoria dependiente de hipocampo en modelos murinos de la enfermedad de Alzheimer Memoria de tesis doctoral presentada por Arnaldo J. Parra Damas para optar al grado de Doctor en Neurociencias por la Universitat Autonòma de Barcelona. Trabajo realizado en la Unidad de Bioquímica y Biología Molecular de la Facultad de Medicina del Departamento de Bioquímica y Biología Molecular de la Universitat Autònoma de Barcelona, y en el Instituto de Neurociencias de la Universitat Autònoma de Barcelona, bajo la dirección del Doctor Carlos Saura Antolín. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Common Genetic Aberrations Associated with Metabolic Interferences in Human Type-2 Diabetes and Acute Myeloid Leukemia: a Bioinformatics Approach
International Journal of Molecular Sciences Article Common Genetic Aberrations Associated with Metabolic Interferences in Human Type-2 Diabetes and Acute Myeloid Leukemia: A Bioinformatics Approach Theodora-Christina Kyriakou 1, Panagiotis Papageorgis 1,2 and Maria-Ioanna Christodoulou 3,* 1 Tumor Microenvironment, Metastasis and Experimental Therapeutics Laboratory, Basic and Translational Cancer Research Center, Department of Life Sciences, European University Cyprus, Nicosia 2404, Cyprus; [email protected] (T.-C.K.); [email protected] (P.P.) 2 European University Cyprus Research Center, Nicosia 2404, Cyprus 3 Tumor Immunology and Biomarkers Laboratory, Basic and Translational Cancer Research Center, Department of Life Sciences, European University Cyprus, Nicosia 2404, Cyprus * Correspondence: [email protected] Abstract: Type-2 diabetes mellitus (T2D) is a chronic metabolic disorder, associated with an increased risk of developing solid tumors and hematological malignancies, including acute myeloid leukemia (AML). However, the genetic background underlying this predisposition remains elusive. We herein aimed at the exploration of the genetic variants, related transcriptomic changes and disturbances in metabolic pathways shared by T2D and AML, utilizing bioinformatics tools and repositories, as well as publicly available clinical datasets. Our approach revealed that rs11709077 and rs1801282, on PPARG, rs11108094 on USP44, rs6685701 on RPS6KA1 and rs7929543 on AC118942.1 comprise Citation: Kyriakou, T.-C.; common SNPs susceptible to the two diseases and, together with 64 other co-inherited proxy SNPs, Papageorgis, P.; Christodoulou, M.-I. may affect the expression patterns of metabolic genes, such as USP44, METAP2, PPARG, TIMP4 and Common Genetic Aberrations RPS6KA1, in adipose tissue, skeletal muscle, liver, pancreas and whole blood. -

Analysis of Chemotherapeutic Response in Ovarian Cancers Using Publicly Available High-Throughput Data
Published OnlineFirst May 21, 2014; DOI: 10.1158/0008-5472.CAN-14-0186 Cancer Therapeutics, Targets, and Chemical Biology Research Analysis of Chemotherapeutic Response in Ovarian Cancers Using Publicly Available High-Throughput Data Jesus Gonzalez Bosquet1, Douglas C. Marchion2, HyeSook Chon3, Johnathan M. Lancaster3, and Stephen Chanock4 Abstract A third of patients with epithelial ovarian cancer (OVCA) will not respond to standard treatment. The determination of a robust signature that predicts chemoresponse could lead to the identification of molecular markers for response as well as possible clinical implementation in the future to identify patients at risk of failing therapy. This pilot study was designed to identify biologic processes affecting candidate pathways associated with chemoresponse and to create a robust gene signature for follow-up studies. After identifying common pathways associated with chemoresponse in serous OVCA in three independent gene-expression experiments, we assessed the biologic processes associated with them using The Cancer Genome Atlas (TCGA) dataset for serous OVCA. We identified differential copy-number alterations (CNA), mutations, DNA meth- ylation, and miRNA expression between patients that responded to standard treatment and those who did not or recurred prematurely. We correlated these significant parameters with gene expression to create a signature of 422 genes associated with chemoresponse. A consensus clustering of this signature identified two differentiated clusters with unique molecular patterns: cluster 1 was significant for cellular signaling and immune response (mainly cell-mediated); and cluster 2 was significant for pathways involving DNA-damage repair and replication, cell cycle, and apoptosis. Validation through consensus clustering was performed in five independent OVCA gene-expression experiments. -
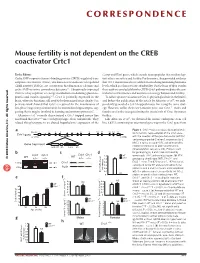
Mouse Fertility Is Not Dependent on the CREB Coactivator Crtc1
CORRESPONDENCE Mouse fertility is not dependent on the CREB coactivator Crtc1 To the Editor: Cartpt and Kiss1 genes, which encode neuropeptides that mediate lep- Cyclic AMP response element–binding protein (CREB)-regulated tran- tin’s effects on satiety and fertility. Furthermore, they provided evidence scription coactivators (Crtcs), also known as transducers of regulated that Crtc1-mutant mice have a deficit in circulating luteinizing hormone CREB activity (TORCs), are coactivators that function as calcium- and levels, which is a characteristic of infertility. On the basis of their results, cyclic AMP–sensitive coincidence detectors1,2. Ubiquitously expressed these authors concluded that the CREB-Crtc1 pathway mediates the cen- Crtc2 is a key regulator of energy metabolism modulating gluconeo- tral effects of hormones and nutrients on energy balance and fertility. genesis and insulin signaling3–5. Crtc1 is primarily expressed in the To follow up our evaluation of Crtc1’s physiological role in the brain6, brain, where its functions still need to be determined more clearly. Our and before the publication of the article by Altarejos et al.7, we inde- previous work showed that Crtc1 is required for the maintenance of pendently generated a Crtc1-trapped mouse line using the same strat- late-phase long-term potentiation in the mammalian hippocampus, sug- egy. However, unlike their Crtc1-mutant mice, our Crtc1–/– males and gesting that it may be involved in learning and memory processes6. females are fertile, thus questioning the crucial role of Crtc1 for mouse Altarejos et al.7 recently characterized a Crtc1-trapped mouse line fertility. and found that Crtc1–/– mice are hyperphagic, obese and infertile. -

CRTC1/MAML2 Gain-Of-Function Interactions with MYC Create A
CRTC1/MAML2 gain-of-function interactions with MYC PNAS PLUS create a gene signature predictive of cancers with CREB–MYC involvement Antonio L. Amelioa,1, Mohammad Fallahib, Franz X. Schauba, Min Zhangc, Mariam B. Lawania, Adam S. Alpersteina, Mark R. Southernd, Brandon M. Younge, Lizi Wuf, Maria Zajac-Kayec,g, Frederic J. Kayec, John L. Clevelanda, and Michael D. Conkrighta,d,2 aDepartment of Cancer Biology, bIT Informatics, dTranslational Research Institute and Department of Molecular Therapeutics, and eGenomics Core, The Scripps Research Institute, Jupiter, FL 33458; and Departments of cMedicine, fMolecular Genetics and Microbiology, and gAnatomy and Cell Biology, Shands Cancer Center, University of Florida, Gainesville, FL 32610 Edited* by Marc Montminy, The Salk Institute for Biological Studies, La Jolla, CA, and approved July 8, 2014 (received for review October 11, 2013) Chimeric oncoproteins created by chromosomal translocations are histology. As such, these are a class of epithelial cell malignancies among the most common genetic mutations associated with tu- that originate from mucous/serous glands present at different morigenesis. Malignant mucoepidermoid salivary gland tumors, as locations in the body (6, 8–15). well as a growing number of solid epithelial-derived tumors, can The genesis of these adult solid tumors was thought to be due arise from a recurrent t (11, 19)(q21;p13.1) translocation that gen- to the direct activation of both CREB and NOTCH/RBPJ, the erates an unusual chimeric cAMP response element binding protein transcription factor targets of the CRTC1 and MAML2 coac- (CREB)-regulated transcriptional coactivator 1 (CRTC1)/mastermind- tivators involved in the t (11, 19) translocation, respectively like 2 (MAML2) (C1/M2) oncoprotein comprised of two transcrip- (6, 15). -

Xo PANEL DNA GENE LIST
xO PANEL DNA GENE LIST ~1700 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes (at 400 -500x average coverage on tumor) Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 -
Comprehensive Pan-Cancer Next Generation Sequencing Solid Tumor Panel, Aberration List Updated 08-07-2020 --Page 1
Comprehensive Pan-Cancer Next Generation Sequencing Solid Tumor Panel, Aberration List Updated 08-07-2020 --Page 1 ABCC3 AR BCL11A CANT1 CDK1 CMKLR1 DAB2IP DUSP9 ABI1 ARAF BCL11B CAPRIN1 CDK12 CNBP DACH1 E2F1 ABL1 ARFRP1 BCL2 CAPZB CDK2 CNOT2 DACH2 E2F3 ABL2 ARHGAP20 BCL2A1 CARD11 CDK4 CNTN1 DAXX EBF1 ABLIM1 ARHGAP26 BCL2L1 CARM1 CDK5RAP2 CNTRL DCLK2 ECT2L ACACA ARHGEF12 BCL2L11 CARS CDK6 COG5 DCN EDIL3 ACE ARHGEF7 BCL2L2 CASC5 CDK7 COL11A1 DDB1 EDNRB ACER1 ARID1A BCL3 CASP3 CDK8 COL1A1 DDB2 EED ACSBG1 ARID1B BCL6 CASP7 CDK9 COL1A2 DDIT3 EEFSEC ACSL3 ARID2 BCL7A CASP8 CDKL5 COL3A1 DDR2 EGF ACSL6 ARID5B BCL9 CAV1 CDKN1A COL6A3 DDX10 EGFR ACVR1 ARIH2 BCOR CBFA2T3 CDKN1B COL9A3 DDX20 EGR1 ACVR1B ARNT BCORL1 CBFB CDKN1C COMMD1 DDX39B EGR2 ACVR1C ARRDC4 BCR CBL CDKN2A COX6C DDX3X EGR3 ACVR2A ASMTL BDNF CBLB CDKN2B CPNE1 DDX41 EGR4 ADD3 ASPH BHLHE22 CBLC CDKN2C CPS1 DDX5 EIF1AX ADM ASPSCR1 BICC1 CCDC28A CDKN2D CPSF6 DDX6 EIF4A2 AFF1 ASTN2 BIN1 CCDC6 CDX1 CRADD DEK EIF4E AFF3 ASXL1 BIRC3 CCDC88C CDX2 CREB1 DGKB ELF3 AFF4 ASXL2 BIRC6 CCK CEBPA CREB3L1 DGKI ELF4 AGR3 ATF1 BLM CCL2 CEBPB CREB3L2 DGKZ ELK4 AHCYL1 ATF3 BMP4 CCNA2 CEBPD CREBBP DICER1 ELL AHI1 ATG13 BMPR1A CCNB1IP1 CEBPE CRKL DIRAS3 ELN AHR ATG5 BRAF CCNB3 CENPF CRLF2 DIS3 ELOVL2 AHRR ATIC BRCA1 CCND1 CENPU CRTC1 DIS3L2 ELP2 AIP ATL1 BRCA2 CCND2 CEP170B CRTC3 DKK1 EML1 AK2 ATM BRCC3 CCND3 CEP57 CSF1 DKK2 EML4 AK5 ATP1B4 BRD1 CCNE1 CEP85L CSF1R DKK4 ENPP2 AKAP12 ATP8A2 BRD3 CCNG1 CHCHD7 CSF3 DLEC1 EP300 AKAP6 ATR BRD4 CCT6B CHD2 CSF3R DLL1 EP400 AKAP9 ATRNL1 BRIP1 CD19 CHD4 CSNK1A1 DLL3 -

CRTC1 Mediates Preferential Transcription at Neuronal Activity- Regulated CRE/TATA Promoters
CRTC1 mediates preferential transcription at neuronal activity- regulated CRE/TATA promoters The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Parra-Damas, Arnaldo, Laura Rubió-Ferrarons, Jie Shen, and Carlos A. Saura. 2017. “CRTC1 mediates preferential transcription at neuronal activity-regulated CRE/TATA promoters.” Scientific Reports 7 (1): 18004. doi:10.1038/s41598-017-18215-y. http:// dx.doi.org/10.1038/s41598-017-18215-y. Published Version doi:10.1038/s41598-017-18215-y Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:34651730 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA www.nature.com/scientificreports OPEN CRTC1 mediates preferential transcription at neuronal activity- regulated CRE/TATA promoters Received: 12 September 2017 Arnaldo Parra-Damas1, Laura Rubió-Ferrarons1, Jie Shen2 & Carlos A. Saura1 Accepted: 27 November 2017 Gene expression mediated by the transcription factor cAMP-responsive element-binding protein Published: xx xx xxxx (CREB) is essential for a wide range of brain processes. The transcriptional coactivartor CREB- regulated transcription coactivator-1 (CRTC1) is required for efcient induction of CREB target genes during neuronal activity. However, the mechanisms regulating induction of specifc CREB/CRTC1- dependent genes during neuronal activity remain largely unclear. Here, we investigated the molecular mechanisms regulating activity-dependent gene transcription upon activation of the CREB/CRTC1 signaling pathway in neurons.