Coffea Arabica L.) Growing in North Sumatra Indonesia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Coffee and Its Effect on Digestion
Expert report Coffee and its effect on digestion By Dr. Carlo La Vecchia, Professor of Medical Statistics and Epidemiology, Dept. of Clinical Sciences and Community Health, Università degli Studi di Milano, Italy. Contents 1 Overview 2 2 Coffee, a diet staple for millions 3 3 What effect can coffee have on the stomach? 4 4 Can coffee trigger heartburn or GORD? 5 5 Is coffee associated with the development of gastric or duodenal ulcers? 6 6 Can coffee help gallbladder or pancreatic function? 7 7 Does coffee consumption have an impact on the lower digestive tract? 8 8 Coffee and gut microbiota — an emerging area of research 9 9 About ISIC 10 10 References 11 www.coffeeandhealth.org May 2020 1 Expert report Coffee and its effect on digestion Overview There have been a number of studies published on coffee and its effect on different areas of digestion; some reporting favourable effects, while other studies report fewer positive effects. This report provides an overview of this body of research, highlighting a number of interesting findings that have emerged to date. Digestion is the breakdown of food and drink, which occurs through the synchronised function of several organs. It is coordinated by the nervous system and a number of different hormones, and can be impacted by a number of external factors. Coffee has been suggested as a trigger for some common digestive complaints from stomach ache and heartburn, through to bowel problems. Research suggests that coffee consumption can stimulate gastric, bile and pancreatic secretions, all of which play important roles in the overall process of digestion1–6. -

Identification of Characterizing Aroma Components of Roasted Chicory
Article Cite This: J. Agric. Food Chem. XXXX, XXX, XXX−XXX pubs.acs.org/JAFC Identification of Characterizing Aroma Components of Roasted Chicory “Coffee” Brews Tiandan Wu and Keith R. Cadwallader* Department of Food Science and Human Nutrition, University of Illinois at Urbana−Champaign, 1302 West Pennsylvania Avenue, Urbana, Illinois 61801, United States *S Supporting Information ABSTRACT: The roasted and ground root of the chicory plant (Cichorium intybus), often referred to as chicory coffee, has served as a coffee surrogate for well over 2 centuries and is still in common use today. Volatile components of roasted chicory brews were identified by direct solvent extraction and solvent-assisted flavor evaporation (SAFE) combined with gas chromatography−olfactometry (GC−O), aroma extract dilution analysis (AEDA), and gas chromatography−mass spectrometry (GC−MS). A total of 46 compounds were quantitated by stable isotope dilution analysis (SIDA) and internal standard methods, and odor-activity values (OAVs) were calculated. On the basis of the combined results of AEDA and OAVs, rotundone was considered to be the most potent odorant in roasted chicory. On the basis of their high OAVs, additional predominant odorants included 3-hydroxy-4,5-dimethyl-2(5H)-furanone (sotolon), 2-methylpropanal, 3-methylbutanal, 2,3- dihydro-5-hydroxy-6-methyl-4H-pyran-4-one (dihydromaltol), 1-octen-3-one, 2-ethyl-3,5-dimethylpyrazine, 4-hydroxy-2,5- dimethyl-3(2H)-furanone (HDMF), and 3-hydroxy-2-methyl-4-pyrone (maltol). Rotundone, with its distinctive aromatic woody, peppery, and “chicory-like” note was also detected in five different commercial ground roasted chicory products. -

Coffee, Coffea Spp
A Horticulture Information article from the Wisconsin Master Gardener website, posted 28 Jan 2013 Coffee, Coffea spp. As you sip your morning cup of coffee have you ever wondered where this ubiquitous beverage comes from? Coffea is a genus of about 100 species of evergreen shrubs and small understory trees in the madder family (Rubiaceae) native to tropical forests in Africa and Asia. The seeds of these plants are processed to produce the drink people around the world have enjoyed for centuries, as well as for fl avoring ice cream, pastries, candies, and liqueurs. It is one of the world’s most valuable crops and is an important export product of several countries. The largest producers include Brazil, Vietnam, Indonesia, and Colombia, along with many other Central and South American countries and East Africa. Coffee comes from a tropical shrub. Coffea is an attractive plant with glossy, deep green foliage. The woody, evergreen shrubs or small trees have opposite, elliptic- ovate, wavy-edged leaves. The fairly stiff leaves have a prominent leaf midrib and lateral veins. Wild plants will grow 10 to 12 feet high, with an open branching structure, but are easily kept smaller and denser by pruning. Fragrant, sweet scented white fl owers bloom along reproductive branches in the leaf axils on old wood. The dense clusters of star-shaped fl owers can be produced at any time of year, but are most common in our Coffea has glossy, deep green leaves. autumn, as coffee is a short-day plant and blooming most profusely when nights are getting longer (daylight of only 8-10 hours). -

Lavazza-Blue-Manual.Pdf
ITALIANO ENGLISH LB2300 FRANÇAIS DEUTSCH ESPAÑOL PORTUGUÊS SINGLE CUP SINGLE CUP WITH STEAM РУССКИЙ DOUBLE CUP WITH STEAM ISTRUZIONI PER L’USO OPERATING INSTRUCTIONS Leggere attentamente queste istruzioni d’uso prima di utilizzare Read these operating instructions carefully before using the la macchina machine BEDIENUNGSANLEITUNG MODE D’EMPLOI Bedienungsanleitung ist vor dem ersten Gebrauch der Maschine Lire attentivement ce mode d’emploi avant d’utiliser la machine aufmerksam zu lesen INSTRUCCIONES PARA EL USO INSTRUÇÕES DE UTILIZAÇÃO Leer atentamente estas instrucciones de uso antes de usar la Leia atentamente estas instruções de utilização antes de utilizar máquina a máquina ИНСТРУКЦИЯ ПО ПРИМЕНЕНИЮ Внимательно прочитайте данную инструкцию перед использованием машины! Type SUP026F - SUP026RF - SUP026RF/US - SUP026RQF - SUP026RQF/US 11 10 4 1 13 2 12 3 9 4 5 6 8 7 15 19 DOUBLE CUP 14 20 WITH STEAM 16 22 17 15 18 21 19 SINGLE CUP 14 23 WITH STEAM 16 22 17 18 21 19 SINGLE CUP 14 23 24 22 17 18 21 • 2 • • 3 • Use of the machine - User Manual IMPORTANT SAFEGUARDS CAUTION When using electrical appliances, basic safety precautions should Any servicing, other than cleaning and user maintenance, should always be followed to reduce the risk of fire, electric shock, and/or be performed by an authorized service centre. Do not immerse injury to persons, including the following: base in water. To reduce the risk of fire or electric shock, do not 1 Read all instructions and information in this instruction book disassemble the base. There are no user serviceable parts inside. and any other literature included in this carton referring to this Repair should be done by authorized service personnel only. -

Coffee, Tea, Or Caffeine-Free?
SPECIAL REPORT: Coffee, Tea, or Caffeine-Free? Copyright 2016 by David L. Meinz, MS, RDN, FAND, CSP www.DavidMeinz.com Americans drink a whopping 500 million cups of coffee every day. That comes to over six billion gallons a year. That’s more than any other country in the world. It’s been our national drink ever since the Boston Tea Party. Coffee accounts for about 75% of the caffeine we take in and about nine out of ten Americans take caffeine in everyday in one form or another. The average American coffee drinker says they take in about 3 ½ cups per day. And the surprising good news about coffee is that there is very little bad news. The coffee bean, like all plants, contains many different naturally occuring compounds and chemicals. Some of those are the good antioxidants that help our body protect itself from damage. As a matter of fact, a recent study found that coffee is the number one source of antioxidants in the U.S; not necessarily because it’s such a good source, but simply because Americans just drink so much of it. It you really want lots of antioxidants, instead of drinking more coffee, start eating more fruit. Blueberries, dates, and red grapes are especially high in antioxidants. Of course the real issue in most peoples minds is the caffeine content of this beverage. There’s no denying that caffeine can improve your mood and help fight fatigue. It can also act as a mild stimulant to improve physical and mental performance especially on monotonous tasks that you do over and over every day. -

The Supply Chain of Fair Trade Coffee: Challenges, Opportunities & the Future Inside a Troubled Industry
University of Vermont ScholarWorks @ UVM Graduate College Dissertations and Theses Dissertations and Theses 2015 The upplS y Chain Of Fair Trade Coffee: Challenges, Opportunities & The uturF e Inside A Troubled Industry Katharine D. Lukas University of Vermont Follow this and additional works at: https://scholarworks.uvm.edu/graddis Part of the Agricultural Economics Commons, and the Sustainability Commons Recommended Citation Lukas, Katharine D., "The uS pply Chain Of Fair Trade Coffee: Challenges, Opportunities & The uturF e Inside A Troubled Industry" (2015). Graduate College Dissertations and Theses. 441. https://scholarworks.uvm.edu/graddis/441 This Thesis is brought to you for free and open access by the Dissertations and Theses at ScholarWorks @ UVM. It has been accepted for inclusion in Graduate College Dissertations and Theses by an authorized administrator of ScholarWorks @ UVM. For more information, please contact [email protected]. THE SUPPLY CHAIN OF FAIR TRADE COFFEE: CHALLENGES, OPPORTUNITIES & THE FUTURE INSIDE A TROUBLED INDUSTRY A Thesis Presented by Katharine Diane Lukas to The Faculty of the Graduate College of The University of Vermont In Partial Fulfillment of the Requirements for the Degree of Master of Science Specializing in Community Development and Applied Economics October, 2015 Defense Date: June 12, 2015 Thesis Examination Committee: David Conner, Ph.D, Advisor Shoshanah Inwood, Ph.D., Chairperson V. Ernesto Méndez, Ph.D. Cynthia J. Forehand, Ph.D., Dean of the Graduate College ABSTRACT What started as a grassroots effort to aid tradespeople in developing nations, Fair Trade and similar certification models have, over the last sixty years, successfully established themselves as a viable alternative to conventional international trade; the ongoing growth of their market share and volume emphasize the increasing market demand for these alternatives. -

Lavazza Roast & Ground Coffees
LAVAZZA ROAST & GROUND COFFEES CAFFÈ ESPRESSO PREMIUM HOUSE BLEND ESPRESSO DECAFFEINATED ¡TIERRA! INTENSO A 100% Arabica blend from A refined blend of 100% A 100% Arabica blend from Central A 100% sustainably grown Arabica Central and South America Arabicas from the highlands of America with full-bodied and sweet, blend. It combines select coffees with highly aromatic notes, Central and South America it long lasting flavor. The natural from South America, providing a fragrant flavor, and a rich delivers an aromatic, fruity and carbon dioxide decaffeination long lasting aroma with chocolate body. Ideal for espresso, fragrant cup of coffee. Ideal for process keeps unaltered its notes. Ideal for espresso, but but also suitable for use in drip coffee preparation. distinctive characteristics. Ideal for also suitable for use in all types of all types of coffee makers. Medium Roast espresso, but also suitable for use coffee makers. Rainforest Alliance Dark Roast in all types of coffee makers. Certified™. Medium Roast Dark Roast CREMA E GUSTO QUALITÀ ORO QUALITÀ ROSSA CAFFÈ DECAFFEINATO A blend that combines Brazilian A selection of the finest 100% Italy’s best-selling coffee A blend of Arabicas from Brazil and Arabicas with African and Arabica coffees from Central since 1971, is a blend high quality Asian Robusta beans. The Indonesian Robusta coffees. An America and the African of Brazilian and African natural carbon dioxide de-caffeination intense and well-balanced aroma, highlands. A blend with a coffees defined by a full process keeps unaltered its distinctive full-bodied and fragrant flavor, fragrant intense aroma, and body, intense aromas and characteristics. -

Chicory, Cichorium Intybus
Did You Know? Chicory, Cichorium intybus ● Common names for chicory include blue sailors, coffeeweed, succory, Italian dandelion, witloof chicory, Belgian endive, French endive. ● Chicory is a perennial in the Aster family with a long tap root and a blue flower. ● The tall flowering stems have blue flowers and produce dandelion-like seeds above low growing foliage. ● Ancient Egyptians grew chicory for medicinal use, a coffee substitute and vegetable crop. ● Traditional medicinal uses varied globally, from malaria treatment in Afghanistan, digestive disorders in Europe, wound healing in Turkey to jaundice in South Africa. The leaves, stem, root and flowers are all used for different preparations. ● The root has up to 40% inulin, which is a type of soluble fiber found in many plants. Chicory root is the main source of inulin used in dietary supplements. It is also used in high fiber, low fat and reduced calorie foods. ● Recent research supports the effectiveness of chicory root a prebiotic, and for improved digestive health and regularity, long term weight management, bone health, blood sugar management and improved metabolic status on blood lipids. ● The dried, roasted, and ground root has long been used as a coffee substitute or coffee extender. ● While ground chicory adds a robust flavor to coffee, it does not contain caffeine. ● Ground chicory has been added to stout beers while brewing for additional flavor. ● In New Orleans, the traditional café au lait is made with rich black coffee, chicory and boiled milk. ● Introduced from Europe, chicory has become naturalized throughout North America. It is listed as a noxious weed in Colorado. -

The Coffee Bean: a Value Chain and Sustainability Initiatives Analysis Melissa Murphy, University of Connecticut, Stamford CT USA Timothy J
The Coffee Bean: A Value Chain and Sustainability Initiatives Analysis Melissa Murphy, University of Connecticut, Stamford CT USA Timothy J. Dowding, University of Connecticut, Stamford CT USA ABSTRACT This paper examines Starbucks’ corporate strategy of sustainable efforts in Ethiopia, particularly in the sustainable sourcing Arabica coffee. The paper discusses the value chain of coffee, issues surrounding the coffee supply chain and the need for sustainable coffee production. In addition it also discusses Starbucks’ position and influence on the coffee trade, and the measures that Starbucks is taking to ensure sustainability efforts throughout the coffee supply chain. COFFEE VALUE CHAIN & P3G ANALYSIS Coffee is produced in more than fifty developing countries in Latin America, Africa, and Asia and it is an important source of income for 20-25 million families worldwide [1]. The initial production of coffee beans including farming, collecting, and processing is labor intensive and as a result is performed in more labor abundant developing countries. The roasting and branding of coffee is more capital intensive and therefore is situated in northern industrialized countries. The top five coffee consumers are United States of America, Brazil, Germany, Japan, and France [1]. The structure of the value chain is very similar regardless of producing or consuming country. The coffee value chain is made up of the four main phases: Cultivation, Processing, Roasting, and Consumption. Each phase in the process has environmental, social, economic -

Coffees Serving Size Caffeine (Mg)
Coffees Serving Size Caffeine (mg) Dunkin' Donuts Coffee with Turbo Shot large, 20 fl. oz. 436 Starbucks Coffee venti, 20 fl. oz. 415 Starbucks Coffee grande, 16 fl. oz. 330 Panera Frozen Mocha 16.5 fl. oz. 267 Starbucks Coffee tall, 12 fl. oz. 260 Starbucks Caffè Americano grande, 16 fl. oz. 225 Panera Coffee regular, 16.8 fl. oz. 189 Starbucks Espresso Frappuccino venti, 24 fl. oz. 185 Dunkin' Donuts Coffee medium, 14 fl. oz. 178 Starbucks Caffè Mocha grande, 16 fl. oz. 175 Starbucks Iced Coffee grande, 16 fl. oz. 165 Maxwell House Ground Coffee—100% Colombian, Dark Roast, 2 Tbs., makes 12 fl. Master Blend, or Original Roast oz. 100-160 Dunkin' Donuts Cappuccino large, 20 fl. oz. 151 Starbucks—Caffè Latte, Cappuccino, or Caramel Macchiato grande, 16 fl. oz. 150 Starbucks Espresso doppio, 2 fl. oz. 150 Keurig Coffee K-Cup, all varieties 1 cup, makes 8 fl. oz. 75-150 2 tsp., makes 12 fl. Folgers Classic Roast Instant Coffee oz. 148 Starbucks Doubleshot Energy Coffee, can 15 fl. oz. 146 Starbucks Mocha Frappuccino venti, 24 fl. oz. 140 1 packet, makes 8 fl. Starbucks VIA House Blend Instant Coffee oz. 135 McDonald's Coffee large, 16 fl. oz. 133 2⅔ Tbs., makes 12- Maxwell House International Café, all flavors 16 fl. oz. 40-130 Seattle's Best Coffee—Iced Latte or Iced Mocha, can 9.5 fl. oz. 90 Starbucks Frappuccino Coffee, bottle 9.5 fl. oz. 90 International Delight Iced Coffee 8 fl. oz. 76 2 Tbs., makes 12 fl. Maxwell House Lite Ground Coffee oz. -

Are Consumers Willing to Pay More for Fair Trade Certified Coffee?
Are Consumers Willing to Pay More for Fair Trade CertifiedTM Coffee? Adam P. Carlson University of Notre Dame Department of Economics [email protected] Instructor: William N. Evans Abstract This study estimates the American consumers’ willingness to pay for Fair Trade Certified coffee using a Hedonic Price Model and Retail Scanner Data. Consistent with prior work, a cross- sectional model finds that Certification has a large positive effect on the price of coffee. However, these models may be subject to an omitted variables bias in that coffees that eventually become Fair Trade Certified are substantially more expensive than other brands. To eliminate this potential bias, I utilize a fixed-effects difference-in-difference specification. Because the certification process takes many months, I observe retail coffee prices for 21 brands before and after a brand becomes Fair Trade Certified. Since the only thing changing over time is certification, I can hold constant the fact that more expensive coffees from the start are more likely to become certified. Comparing this time series change in prices with the changes that occur over time for other coffees, this model finds Certification to cause a minor increase in the price of coffee of about $0.12, a change of 1.1 percent, -- a substantially smaller estimate than has been found in the past. These results suggest that consumers are willing to pay only a small premium for Fair Trade coffee. I. Introduction Traditional models in economics assume that the price of a good is determined by the desirability of the physical characteristics of that good. -
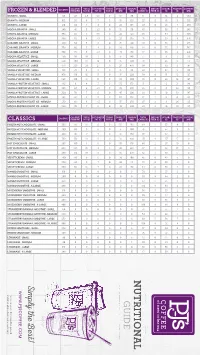
Pjsnutritionalinfo.Pdf
CALORIES TOTAL SATURATED TRANS CHOLEST. SODIUM TOTAL DIETARY SUGARS PROTEIN CAFFEINE FROZEN & BLENDED CALORIES FROM FAT FAT (G) FAT (G) FAT (G) (MG) (MG) CARBS (G) FIBER (G) (G) (G) (MG) GRANITA - SMALL 120 40 4.5 2.5 0 10 80 17 0 14 4 294 GRANITA - MEDIUM 160 50 6 3 0 15 105 23 0 19 5 392 GRANITA - LARGE 210 60 7 4 0 20 135 29 0 23 7 490 MOCHA GRANITA - SMALL 240 45 5 2.5 0 15 90 43 1 36 5 245 MOCHA GRANITA - MEDIUM 350 60 7 3.5 0 20 120 65 2 54 7 343 MOCHA GRANITA - LARGE 390 70 8 4.5 0 25 150 71 2 59 8 441 CARAMEL GRANITA - SMALL 240 50 5 3 0 15 120 43 0 38 4 245 CARAMEL GRANITA - MEDIUM 330 60 7 4 0 20 140 61 0 55 5 343 CARAMEL GRANITA - LARGE 390 70 8 4.5 0 25 170 72 0 65 6 441 MOCHA VELVET ICE - SMALL 250 90 10 7 0 15 140 37 1 31 3 98 MOCHA VELVET ICE - MEDIUM 530 200 22 18 0 15 360 78 1 65 5 123 MOCHA VELVET ICE - LARGE 670 260 29 23 0 20 470 99 1 82 6 147 VANILLA VELVET ICE - SMALL 190 70 8 6 0 15 90 22 0 20 5 98 VANILLA VELVET ICE - MEDIUM 430 150 16 13 0 15 220 58 0 51 12 123 VANILLA VELVET ICE - LARGE 560 190 21 17 0 20 300 76 0 66 16 147 VANILLA PROTEIN VELVET ICE - SMALL 190 9 4 3 0 30 170 15 1 6 21 98 VANILLA PROTEIN VELVET ICE - MEDIUM 250 60 6 4.5 0 35 230 22 2 8 26 123 VANILLA PROTEIN VELVET ICE - LARGE 320 70 7 6 0 45 290 28 3 10 32 147 MOCHA PROTEIN VELVET ICE - SMALL 190 40 4 3 0 30 170 17 1 6 21 98 MOCHA PROTEIN VELVET ICE - MEDIUM 250 60 6 4.5 0 35 230 23 2 8 26 123 MOCHA PROTEIN VELVET ICE - LARGE 320 70 7 6 0 45 290 29 2 10 32 147 CALORIES TOTAL SATURATED TRANS CHOLEST.