Identification of Claudin 1 Transcript Variants in Human Invasive Breast Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Analysis of the Indacaterol-Regulated Transcriptome in Human Airway
Supplemental material to this article can be found at: http://jpet.aspetjournals.org/content/suppl/2018/04/13/jpet.118.249292.DC1 1521-0103/366/1/220–236$35.00 https://doi.org/10.1124/jpet.118.249292 THE JOURNAL OF PHARMACOLOGY AND EXPERIMENTAL THERAPEUTICS J Pharmacol Exp Ther 366:220–236, July 2018 Copyright ª 2018 by The American Society for Pharmacology and Experimental Therapeutics Analysis of the Indacaterol-Regulated Transcriptome in Human Airway Epithelial Cells Implicates Gene Expression Changes in the s Adverse and Therapeutic Effects of b2-Adrenoceptor Agonists Dong Yan, Omar Hamed, Taruna Joshi,1 Mahmoud M. Mostafa, Kyla C. Jamieson, Radhika Joshi, Robert Newton, and Mark A. Giembycz Departments of Physiology and Pharmacology (D.Y., O.H., T.J., K.C.J., R.J., M.A.G.) and Cell Biology and Anatomy (M.M.M., R.N.), Snyder Institute for Chronic Diseases, Cumming School of Medicine, University of Calgary, Calgary, Alberta, Canada Received March 22, 2018; accepted April 11, 2018 Downloaded from ABSTRACT The contribution of gene expression changes to the adverse and activity, and positive regulation of neutrophil chemotaxis. The therapeutic effects of b2-adrenoceptor agonists in asthma was general enriched GO term extracellular space was also associ- investigated using human airway epithelial cells as a therapeu- ated with indacaterol-induced genes, and many of those, in- tically relevant target. Operational model-fitting established that cluding CRISPLD2, DMBT1, GAS1, and SOCS3, have putative jpet.aspetjournals.org the long-acting b2-adrenoceptor agonists (LABA) indacaterol, anti-inflammatory, antibacterial, and/or antiviral activity. Numer- salmeterol, formoterol, and picumeterol were full agonists on ous indacaterol-regulated genes were also induced or repressed BEAS-2B cells transfected with a cAMP-response element in BEAS-2B cells and human primary bronchial epithelial cells by reporter but differed in efficacy (indacaterol $ formoterol . -

Claudin-1, -3 and -4 Proteins and Mrna Expression in Benign and Malignant Breast Lesions: a Research Study
Available online http://breast-cancer-research.com/content/7/2/R296 ResearchVol 7 No 2 article Open Access Claudin-1, -3 and -4 proteins and mRNA expression in benign and malignant breast lesions: a research study Anna-Mária Tőkés1*, Janina Kulka1*, Sándor Paku2, Ágnes Szik1, Csilla Páska1, Pál Kaposi Novák1, László Szilák1, András Kiss1, Krisztina Bögi1 and Zsuzsa Schaff1 12nd Department of Pathology, Semmelweis University, Budapest, Hungary 2Department of Molecular Pathology, Joint Research Organization of the Hungarian Academy of Sciences, Budapest, Hungary * Contributed equally Corresponding author: Janina Kulka, [email protected] Received: 23 Jan 2004 Revisions requested: 15 Mar 2004 Revisions received: 1 Oct 2004 Accepted: 2 Dec 2004 Published: 31 Jan 2005 Breast Cancer Research 2005, 7:R296-R305 (DOI 10.1186/bcr983)http://breast-cancer-research.com/content/7/2/R296 © 2005 Tőkés et al., licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/ 2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is cited. Abstract Introduction We compared levels of protein and mRNA CLDN4 was present in all 56 tissue sections. However, CLDN4 expression of three members of the claudin (CLDN) family in was highly positive in normal epithelial cells and was decreased malignant breast tumours and benign lesions. or absent in 17 out of 21 ductal carcinoma grade 1, in special types of breast carcinoma (mucinous, papillary, tubular) and in Methods Altogether, 56 sections from 52 surgically resected areas of apocrine metaplasia. -

Identification of Potential Core Genes in Sevoflurance Induced Myocardial
Identication of Potential core genes in Sevourance induced Myocardial Energy Metabolism in Patients Undergoing Off-pump Coronary Artery Bypass Graft Surgery using Bioinformatics analysis Hua Lin ( [email protected] ) Tianjin Medical University General Hospital Airport Site Research article Keywords: sevourane, Myocardial Energy Metabolism, Off-pump Coronary Artery Bypass Graft Surgery Posted Date: November 18th, 2019 DOI: https://doi.org/10.21203/rs.2.17434/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/15 Abstract Background: Myocardial ischemia-reperfusion injury always happened after Off-pump coronary artery bypass graft(OPCABG), and this can not be avoided altogether. In this study, we tried to detect potential genes of sevourane-induced myocardial energy metabolism in patients undergoing OPCABG using bioinformatics analysis. Methods: We download and analyze the gene expression prole data from the Gene Expression Omnibus(GEO) database using bioinformatics methods. We downloded the gene expression data from the Gene Expression Omnibus(GEO) database using bioinformatics methods. Gene Ontology(GO) functional annotation analysis and Kyoto Encyclopedia of Genes and Genomes(KEGG) pathway enrichment analysis were used to analysis the screened differentially expressed genes(DEGs). Then, we established a protein–protein interaction (PPI) network to nd hub genes associated with myocardial energy metabolism. Results: Through PPI network, we nd ten hub genes, including JUN, EGR1, ATF3, FOSB, JUNB, DUSP1, EGR2, NR4A1, BTG2, NR4A2. Conclusions: In conclusion, the proteins encoded by EGR1ATF3c-FosBtg2JunBDUSP1NR4A1BTG2 and NR4A2 were related to cardiac function. ATF3, FOSB, JUNB, DUSP1, NR4A1, NR4A2 are related to apoptosis of cardiomyocytes. The protein encoded by BTG2 is related to hypertrophy. -

Claudin-1, a Double-Edged Sword in Cancer
International Journal of Molecular Sciences Review Claudin-1, A Double-Edged Sword in Cancer Ajaz A. Bhat 1 , Najeeb Syed 1, Lubna Therachiyil 2,3, Sabah Nisar 1, Sheema Hashem 1, Muzafar A. Macha 4,5, Santosh K. Yadav 1, Roopesh Krishnankutty 2, Shanmugakonar Muralitharan 6, Hamda Al-Naemi 6, Puneet Bagga 7, Ravinder Reddy 7, Punita Dhawan 5, Anthony Akobeng 8, Shahab Uddin 2 , Michael P. Frenneaux 9, Wael El-Rifai 10 and Mohammad Haris 1,6,* 1 Division of Translational Medicine, Research Branch, Sidra Medicine, Doha 26999, Qatar; [email protected] (A.A.B.); [email protected] (N.S.); [email protected] (S.N.); [email protected] (S.H.); [email protected] (S.K.Y.) 2 Translational Research Institute, Academic Health System, Hamad Medical Corporation, Doha 3050, Qatar; [email protected] (L.T.); [email protected] (R.K.); [email protected] (S.U.) 3 Department of Pharmaceutical Sciences, College of Pharmacy, QU Health, Qatar University, Doha 2713, Qatar 4 Department of Biotechnology, Central University of Kashmir, Ganderbal, Jammu and Kashmir 191201, India; [email protected] 5 Department of Biochemistry and Molecular Biology, University of Nebraska Medical Center, Omaha, NE 68198, USA; [email protected] 6 Laboratory Animal Research Center, Qatar University, Doha 2713, Qatar; [email protected] (S.M.); [email protected] (H.A.-N.) 7 Center for Magnetic Resonance and Optical Imaging, Department of Radiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104, USA; [email protected] -

Antibody Tools Immunohistochemistry
Antibody Tools Immunohistochemistry $$ 250 - 150 - 100 - 75 - 50 - 37 - Western Blot 25 - 20 - 15 - 10 - 1.4 1.2 1 0.8 0.6 OD 450 0.4 Sandwich ELISA 0.2 0 0.01 0.1 1 10 100 1000 Recombinant Protein Concentration(mg/ml) Immunohistochemistry Immunofluorescence 1 2 3 250 - 150 - 100 - 75 - 50 - Immunoprecipitation 37 - 25 - 20 - 15 - 100 80 60 % of Max 40 Flow Cytometry 20 0 3 4 5 0 102 10 10 10 www.abnova.com March 2013 (Sixth Edition) Abnova Corporation www.abnova.com Email: [email protected] Address: 9F, No. 108, Jhouzih St., Neihu, Taipei 114, Taiwan Tel: + 886 2 8751 1888 Fax: + 886 2 6602 1218 Antibodies tool for IHC, Class I IVD (In Vitro Diagnostics) Cat. Num. DH0003 DH0013 DH0020 DH0015 Product Name Anti-ACTN4 monoclonal antibody Anti-ANXA5 monoclonal antibody Anti-CDH17 monoclonal antibody Anti-CLDN1 monoclonal antibody Application Immunoperoxidase of monoclonal antibody Immunoperoxidase of monoclonal antibody to Immunoperoxidase of monoclonal antibody to Immunoperoxidase of monoclonal antibody to to ACTN4 on formalin-fixed paraffin- ANXA5 on formalin-fixed paraffin-embedded CDH17 on formalin-fixed paraffin-embedded CLDN1 on formalin-fixed paraffin-embedded embedded human pancreatic cancer. [antibody human colon cancer. [antibody concentration 3 human colon cancer. [antibody concentration 3 human colon cancer. [antibody concentration 3 concentration 1.5 ug/ml] ug/ml] ug/ml] ug/ml] Cat. Num. DH0002 DH0021 DH0010 DH0017 Product Name Anti-CTH monoclonal antibody Anti-EGR1 monoclonal antibody Anti-EIF2C2 monoclonal antibody Anti-ENO1 monoclonal antibody Application Immunoperoxidase of monoclonal antibody Immunoperoxidase of monoclonal antibody Immunoperoxidase of monoclonal antibody to Immunoperoxidase of monoclonal antibody to CTH on formalin-fixed paraffin-embedded to EGR1 on formalin-fixed paraffin-embedded EIF2C2 on formalin-fixed paraffin-embedded to ENO1 on formalin-fixed paraffin-embedded human liver. -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN. -

Chromosome Transfer Induced Aneuploidy Results in Complex Dysregulation of the Cellular Transcriptome in Immortalized and Cancer Cells
[CANCER RESEARCH 64, 6941–6949, October 1, 2004] Chromosome Transfer Induced Aneuploidy Results in Complex Dysregulation of the Cellular Transcriptome in Immortalized and Cancer Cells Madhvi B. Upender,1 Jens K. Habermann,1,4 Lisa M. McShane,3 Edward L. Korn,3 J. Carl Barrett,2 Michael J. Difilippantonio,1 and Thomas Ried1 1Genetics Branch and 2Laboratory for Biosystems and Cancer, Center for Cancer Research and 3Biometric Research Branch, National Cancer Institute/NIH, Bethesda, Maryland; and 4Department of Oncology and Pathology, Cancer Center Karolinska, Karolinska Institute, Stockholm, Sweden ABSTRACT tumor cells (4, 18–20). Additionally, in cell culture model systems in which cells are exposed to different carcinogens, chromosomal ane- Chromosomal aneuploidies are observed in essentially all sporadic uploidy is the earliest detectable genomic aberration (21, 22). carcinomas. These aneuploidies result in tumor-specific patterns of The conservation of these tumor and tumor-stage–specific patterns genomic imbalances that are acquired early during tumorigenesis, con- tinuously selected for and faithfully maintained in cancer cells. Although of chromosomal aneuploidies suggests that they play a fundamental the paradigm of translocation induced oncogene activation in hematologic biological role in tumorigenesis. It remains, however, unresolved how malignancies is firmly established, it is not known how genomic imbal- such genomic imbalances affect global gene expression patterns. One ances affect chromosome-specific gene expression patterns in particular could postulate that expression levels of all transcriptionally active and how chromosomal aneuploidy dysregulates the genetic equilibrium of genes on trisomic chromosomes would increase in accordance with cells in general. To model specific chromosomal aneuploidies in cancer the chromosome copy number. -

Primary Gene Structure and Expression Studies of Rodent Paracellin-1
J Am Soc Nephrol 12: 2664–2672, 2001 Primary Gene Structure and Expression Studies of Rodent Paracellin-1 STEFANIE WEBER,* KARL P. SCHLINGMANN,* MELANIE PETERS,* LENE NIEMANN NEJSUM,† SØREN NIELSEN,† HARTMUT ENGEL,‡ KARL-HEINZ GRZESCHIK,‡ HANNSJO¨ RG W. SEYBERTH,* HERMANN-JOSEPH GRO¨ NE,§ ROLF NU¨ SING,* and MARTIN KONRAD* *Department of Pediatrics, Philipps University, Marburg, Germany; †Department of Cell Biology, Institute of Anatomy, University of Aarhus, Aarhus, Denmark; ‡Institute of Human Genetics, Philipps University, Marburg, Germany; and §Deutsches Krebsforschungszentrum, Heidelberg, Germany. Abstract. The novel member of the claudin multigene family, lengths and splice-site loci. By radiation hybrid mapping, the paracellin-1/claudin-16, encoded by the gene PCLN1, is a renal murine Pcln1 gene was assigned directly to marker D16Mit133 tight junction protein that is involved in the paracellular trans- on mouse chromosome 16 (syntenic to a locus on human port of magnesium and calcium in the thick ascending limb of chromosome 3q27, which harbors the human PCLN1 gene). Henle’s loop. Mutations in human PCLN1 are associated with Mouse multiple-tissue Northern blot showed Pcln1 expression familial hypomagnesemia with hypercalciuria and nephrocal- exclusively in the kidney. The expression profile along the cinosis, an autosomal recessive disease that is characterized by nephron was analyzed by reverse transcriptase–PCR on micro- severe renal magnesium and calcium loss. The complete cod- dissected nephron segments and immunohistochemistry of rat ing sequences of mouse and rat Pcln1 and the murine genomic kidney. Paracellin-1 expression was restricted to distal tubular structure are here presented. Full-length cDNAs are 939 and segments including the thick ascending limb of Henle’s loop, 1514 bp in length in mouse and rat, respectively, encoding a the distal tubule, and the collecting duct. -

Identification of Claudin‑1, ‑3, ‑7 and ‑8 As Prognostic Markers in Human Laryngeal Carcinoma
MOLECULAR MEDICINE REPORTS 20: 393-400, 2019 Identification of claudin‑1, ‑3, ‑7 and ‑8 as prognostic markers in human laryngeal carcinoma SHU ZHOU1, XUE PIAO2, CHENGYAN WANG3, RUI WANG3 and ZHIMIN SONG4 1Department of Anesthesiology, Jilin Cancer Hospital; 2Department of Anesthesiology, Maternity Hospital of Changchun City; 3Department of Ultrasound, Jilin Cancer Hospital; 4Department of Anesthesiology, The Second Hospital of Jilin University, Changchun, Jilin 130021, P.R. China Received June 20, 2018; Accepted February 8, 2019 DOI: 10.3892/mmr.2019.10265 Abstract. Various genomic and epigenetic modifications demonstrated that the expression levels of CLDN1, -3, -7 and that occur during the development of cancer act as potential -8 varied between laryngeal squamous carcinoma tissues and biomarkers for early diagnosis and treatment. Previous studies non-neoplastic tissues. The expression levels of these CLDNs have demonstrated abnormal expression of the claudin (CLDN) may be useful molecular markers for the diagnosis of laryngeal tight junction (TJ) proteins in numerous types of human carcinoma, and determining the metastasis and prognosis of this cancer. Reverse transcription-quantitative polymerase chain disease. reaction and western blotting were employed to investigate variations in the expression of the CLDN TJ proteins in laryn- Introduction geal non-neoplastic tissues and laryngeal squamous carcinoma tissues. It was revealed that CLDN2, CLDN4, CLDN5, CLDN6, Tight junctions (TJs) form the apical junctional complex in epithe- CLDN9, CLDN11 and CLDN12 were undetectable in laryngeal lial and endothelial cellular sheets in cooperation with adherens squamous carcinoma tissues and laryngeal non-neoplastic junctions and desmosomes (1). TJs are important for the tight tissues. Additionally, CLDN10 was expressed in laryngeal squa- structure of cellular sheets, which enables monitored paracel- mous carcinoma tissues and laryngeal non-neoplastic tissues; lular ion flux and the maintenance of tissue homeostasis (2). -

Us 2018 / 0305689 A1
US 20180305689A1 ( 19 ) United States (12 ) Patent Application Publication ( 10) Pub . No. : US 2018 /0305689 A1 Sætrom et al. ( 43 ) Pub . Date: Oct. 25 , 2018 ( 54 ) SARNA COMPOSITIONS AND METHODS OF plication No . 62 /150 , 895 , filed on Apr. 22 , 2015 , USE provisional application No . 62/ 150 ,904 , filed on Apr. 22 , 2015 , provisional application No. 62 / 150 , 908 , (71 ) Applicant: MINA THERAPEUTICS LIMITED , filed on Apr. 22 , 2015 , provisional application No. LONDON (GB ) 62 / 150 , 900 , filed on Apr. 22 , 2015 . (72 ) Inventors : Pål Sætrom , Trondheim (NO ) ; Endre Publication Classification Bakken Stovner , Trondheim (NO ) (51 ) Int . CI. C12N 15 / 113 (2006 .01 ) (21 ) Appl. No. : 15 /568 , 046 (52 ) U . S . CI. (22 ) PCT Filed : Apr. 21 , 2016 CPC .. .. .. C12N 15 / 113 ( 2013 .01 ) ; C12N 2310 / 34 ( 2013. 01 ) ; C12N 2310 /14 (2013 . 01 ) ; C12N ( 86 ) PCT No .: PCT/ GB2016 /051116 2310 / 11 (2013 .01 ) $ 371 ( c ) ( 1 ) , ( 2 ) Date : Oct . 20 , 2017 (57 ) ABSTRACT The invention relates to oligonucleotides , e . g . , saRNAS Related U . S . Application Data useful in upregulating the expression of a target gene and (60 ) Provisional application No . 62 / 150 ,892 , filed on Apr. therapeutic compositions comprising such oligonucleotides . 22 , 2015 , provisional application No . 62 / 150 ,893 , Methods of using the oligonucleotides and the therapeutic filed on Apr. 22 , 2015 , provisional application No . compositions are also provided . 62 / 150 ,897 , filed on Apr. 22 , 2015 , provisional ap Specification includes a Sequence Listing . SARNA sense strand (Fessenger 3 ' SARNA antisense strand (Guide ) Mathew, Si Target antisense RNA transcript, e . g . NAT Target Coding strand Gene Transcription start site ( T55 ) TY{ { ? ? Targeted Target transcript , e . -
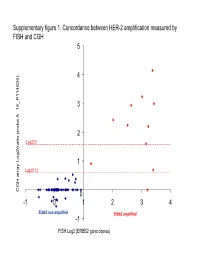
Supplementary Data
Supplementary figure 1. Concordance between HER-2 amplification measured by FISH and CGH 5 4 6) 3 14_P11482 __ 2 Log2(3) atio probe A atio probe rr 1 Log2(1.5) array Log2( CGH 0 -1 0 1 2 3 4 Er bb2 non amplifi e d Erbb2 amplified -1 FISH Log2 (ERBB2 gene copies) Supplementary Figure 2. Gained and lost regions according to triple-negativity Non-triple negative tumors (n=69) Triple negative tumors (n=35) Non-triple negative tumors (n=69) vs triple negative tumors (n=35) Supplementary Figure 3. Gained and lost regions according to chemotherapy response Supplementary Figure 4. Gained and lost regions according to taxanes efficacy Supplementary Table 1. Patient characteristics (n=106). Characteristic N (%) Age (median, range) 51 (28-73) cT cT0/T1 6 (6%) cT2 53 (50%) cT3 18 (17%) cT4 29 (17%) cN cN0 24 (23%) cN1 50 (47%) cN2 23 (22%) cN3 9 (8%) Tumor grade Grade 1 6 (6%) Grade 2 39 (37%) Grade 3 47 (43%) Not assessable 14 (14%) Estrogen receptor expression Negative 45 (42%) Positive 60 (57%) Unknown 1 (1%) Progesteron receptor expression Negative 57 (54%) Positive 47 (44%) Unknown 2 (2%) Her2 status Normal 95 (90%) Overexpression and/or amplification 11 (10%) Preoperative chemotherapy T/FAC* 66 (62%) FAC 38 (36%) Trastuzumab-containing regimen 2 (2%) Pathologic complete remission No 83 (78%) Yes 20 (19%) Not assessable 3 (3%) *: Sequential Paclitaxel / FAC (or FEC) or docetaxel /FEC (n=2) Supplementary Table 2. Minimum Common region (MCR) gained in more than 20% of samples: Band1 Probes Freq Genes AP2M1 CLCN2 DVL3 EIF4G1 EPHB3 POLR2H PSMD2 THPO