Genetic Studies in a Tropical Pine - Pinus Kesiya II
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phytogeographic Review of Vietnam and Adjacent Areas of Eastern Indochina L
KOMAROVIA (2003) 3: 1–83 Saint Petersburg Phytogeographic review of Vietnam and adjacent areas of Eastern Indochina L. V. Averyanov, Phan Ke Loc, Nguyen Tien Hiep, D. K. Harder Leonid V. Averyanov, Herbarium, Komarov Botanical Institute of the Russian Academy of Sciences, Prof. Popov str. 2, Saint Petersburg 197376, Russia E-mail: [email protected], [email protected] Phan Ke Loc, Department of Botany, Viet Nam National University, Hanoi, Viet Nam. E-mail: [email protected] Nguyen Tien Hiep, Institute of Ecology and Biological Resources of the National Centre for Natural Sciences and Technology of Viet Nam, Nghia Do, Cau Giay, Hanoi, Viet Nam. E-mail: [email protected] Dan K. Harder, Arboretum, University of California Santa Cruz, 1156 High Street, Santa Cruz, California 95064, U.S.A. E-mail: [email protected] The main phytogeographic regions within the eastern part of the Indochinese Peninsula are delimited on the basis of analysis of recent literature on geology, geomorphology and climatology of the region, as well as numerous recent literature information on phytogeography, flora and vegetation. The following six phytogeographic regions (at the rank of floristic province) are distinguished and outlined within eastern Indochina: Sikang-Yunnan Province, South Chinese Province, North Indochinese Province, Central Annamese Province, South Annamese Province and South Indochinese Province. Short descriptions of these floristic units are given along with analysis of their floristic relationships. Special floristic analysis and consideration are given to the Orchidaceae as the largest well-studied representative of the Indochinese flora. 1. Background The Socialist Republic of Vietnam, comprising the largest area in the eastern part of the Indochinese Peninsula, is situated along the southeastern margin of the Peninsula. -

Chapter 1 Introduction
Chapter 1 Introduction Chapterl Page 1 1.1 Introduction Chir pine scientifically known as Pinus roxburghii (family Coniferae) is one of the six Pinus species of India and the most widely occurring. It is also known as Himalayan long needle pine, long leaved Indian pine, Indian chir pine, chir or chil. Chir pine (Pinus roxburghii), is the dominant species native of the inter-ranges and principal valleys of the Himalaya. It begins from Afghanistan in the west and ends in Bhutan in the east and extends through countries like Pakistan, India and Nepal. In India its natural forests are found in Jammu and Kashmir, Haryana, Uttarakhand, Himachal Pradesh, and Uttar Pradesh, parts of Sikkim, West Bengal and Arunachal Pradesh. The total area under chir forests is estimated to be 8, 90,000 hectares which occurs between 450 m to 2300 m altitude. It forms pure forests in its habitat but in its upper and lower limits occurs mixed with other conifers and broad leaved species, though rarely, in its upper limits it is found in association with deodar, kail, ban oak, burans (Rhododendron) etc., and in the lower limits with sal, sain, khair, harada, bahera, amla, jamun etc. Besides Chir pine other indigenous pines which are present in Indian subcontinent are P. gerardiana, P. wallichiana, P.kesiya, P.bhutanica, and P.merkusii (Ginwal et.al, 2009), all belong to family Abitaceae or Pinaceae of order coniferals. Classification of Genus Pinus : Kingdom : Plantae Division : Pinophyta Class : Pinopsida Order : Pinales / Coniferals Family : Pinacea/Abitaceae Genus : Pinus The pines have been traced back in the geological history to Jurassic period (150 million years) reaching their climax in the tertiary period (60 million years). -

Pinus Kesiya Royle Ex Gord
Indian Journal of Biotechnology Vol 6, April 2007, pp 262-266 Plantlet regeneration of Pinus kesiya Royle ex Gord. from mature embryos Pramod Tandon*, Suman Kumaria and Hiranjit Choudhury Plant Biotechnology Laboratory, Department of Botany, North-Eastern Hill University, Shillong 793 022, India Received 7 September 2005; revised 8 May 2006; accepted 10 August 2006 A high frequency (90.5%) of shoot bud induction was observed in mature zygotic embryos of Pinus kesiya Royle ex Gord. that were pre-cultured in Quoirin and Lepoivre (LP) medium containing 23.15 µM Kn for 4 wk and then transferred to growth regulator-free medium. Multiplication and elongation of the shoot buds resulted in 1/2 LP medium containing 0.5 µM IBA and 4.5 µM BAP or Kn. Rooting was 72.3% in isolated shoots that were treated with 53.76 µΜ ΝΑΑ for 24 h and then cultured on water-agar medium. Plantlets were hardened and successfully established on soil collected from pine forests with 70% survival. Biomass of the micropropagated plants was 3.2 times higher than seed- derived plants. Keywords : growth characteristics, micropropagation, shoot buds induction, zygotic embryos, Pinus kesiya IPC Code: Int. Cl. 8 A01H4/00, 7/00 Introduction were surface-sterilized with 1% (w/v) mercuric Pinus kesiya Royle ex Gord. (Khasi pine) is chloride for 2-3 min, washed 3-4 times with sterilized widely distributed in the eastern Himalayan region of pure water and then treated with 6% (v/v) hydrogen the Indian subcontinent and extends up to Myanmar, peroxide for 10 min and stratified at 4°C for 24 h. -

Seasonal Abundance and Diversity of Arthropods on Acacia Mangium (Fabales: Fabaceae) Trees As Windbreaks in the Cerrado F
Seasonal abundance and diversity of arthropods on Acacia mangium (Fabales: Fabaceae) trees as windbreaks in the cerrado F. W. S. Silva1,2,*, G. L. D. Leite1, R. E. M. Guañabens1, R. A. Sampaio1, C. A. G. Gusmão1, J. E. Serrão3 and J. C. Zanuncio4 Abstract Our aim was to assess the seasonal abundance and diversity of arthropods on Acacia mangium Willd. (Fabales: Fabaceae) trees. Phytophagous arthropods, natural enemies and pollinators were quantified biweekly in 20 trees during 3 years. The Shannon biodiversity index (H’) of arthro- pods was greater in the summer and smaller in the winter and spring on A. magium. The diversities of species and abundances of individuals of all arthropod taxa were greater in the autumn and smaller in the winter and in the spring. The H’ index of arthropods was greater in 2005 and smaller in 2007. The diversity of species and abundance of individuals of phytophagous taxa were greater in 2005 and 2006, respectively, whereas those of natural enemies and pollinators was greater in 2006. Aethalion reticulatum L. (Hemiptera: Aetalionidae) was the most abundant phytophagous species found in the various seasons, while Camponotus sp.2 (Hymenoptera: Formicidae), Trigona spinipes Fabricius (Hymenoptera: Apidae, Meliponini), Tetragonisca angustula Latreille (Hymenoptera: Meliponinae, Meliponini) and Polistes sp. (Hymenoptera: Vespidae) were the most abundant natural enemies and pollinators. The great diversity of predators on this tree species can explain the little damage to its leaves. These results may be applied to support programs of pest control and maintenance of natural enemies and pollinators in future plantations of A. mangium. Key Words: biodiversity; insects, phytophagous; natural enemies and pollinators Resumen Nuestro objetivo era evaluar la abundancia y diversidad estacional de artrópodos en Acacia mangium Willd. -

PINUS KESIYA Page 1Of 4
PINUS KESIYA Page 1of 4 Family: PINACEAE (gymnosperm) Scientific name(s): Pinus kesiya Commercial restriction: no commercial restriction Note: This species can be found at altitudes between 600 m and 2700 m. WOOD DESCRIPTION LOG DESCRIPTION Color: orange - yellow Diameter: from 50 to 60 cm Sapwood: clearly demarcated Thickness of sapwood: from 4 to 5 cm Texture: medium Floats: yes Grain: straight Log durability: low (must be treated) Interlocked grain: absent Note: Wood pinkish white. Numerous resin canals. PHYSICAL PROPERTIES MECHANICAL AND ACOUSTIC PROPERTIES Physical and mechanical properties are based on mature heartwood specimens. These properties can vary greatly depending on origin and growth conditions. Mean Std dev. Mean Std dev. Specific gravity *: 0,71 Crushing strength *: 65 MPa Monnin hardness *: 2,8 Static bending strength *: 85 MPa Coeff. of volumetric shrinkage: 0,52 % Modulus of elasticity *: 12300 MPa Total tangential shrinkage (TS): 8,9 % Total radial shrinkage (RS): 6,1 % (*: at 12% moisture content, with 1 MPa = 1 N/mm²) TS/RS ratio: 1,5 Fiber saturation point: 35 % Musical quality factor: 96,2 measured at 2718 Hz Stability: moderately stable Note: Physical and mechanical properties vary according to the age and the origin. NATURAL DURABILITY AND TREATABILITY Fungi and termite resistance refers to end-uses under temperate climate. Except for special comments on sapwood, natural durability is based on mature heartwood. Sapwood must always be considered as non-durable against wood degrading agents. E.N. = Euro Norm Funghi (according to E.N. standards): class 4 - poorly durable Dry wood borers: susceptible Termites (according to E.N. standards): class S - susceptible Treatability (according to E.N. -

Pinus Kesiya Royle Ex
Pinus kesiya Royle ex. Gordon Schmidt, Lars Holger; Nguyen, Duc To Luu Published in: Seed Leaflet Publication date: 2004 Document version Publisher's PDF, also known as Version of record Citation for published version (APA): Schmidt, L. H., & Nguyen, D. T. L. (2004). Pinus kesiya Royle ex. Gordon. Seed Leaflet, (93). Download date: 29. Sep. 2021 SEED LEAFLET No. 93 December 2004 VTSP Pinus kesiya Royle ex. Gordon Taxonomy and nomenclature Family: Pinaceae Synonyms: Pinus khasya Hook f., Pinus insularis Endl., Vernacular/common names: khasiya pine, khasi pine (Eng.); benguet pine (Philippines); pin a trois feuilles (France); son-sambai (Thailand); thong ba la (Vietnam). Note: the taxonomic delineation of P. kesiya is disputed as it appears morphologically indistin- guishable from P. yunnanensis Franch. Distribution and habitat Range of distribution covers large area of SE Asia in- cluding Thailand, Burma, Laos, S. China and Vietnam Female cones and male strobili at the time of pollination. to the Philippines. Normally confi ned to altitudes 700 Photo: Lars Schmidt -1200 m.a.s.l., in east India however, reported up to 3000 m. Often forming pure stands. When growing in the same area as P. merkusii the two species may Fruit and seed description be co-dominant in an overlapping altitude with P. Fruit: dehiscent cones, ovoid, sessile, 5-9 cm long, merkusii being more frequent at lower altitudes and P. 3-5 cm in diameter, cone scales dense. Old cones dark kesiya at higher. At higher altitudes occasionally co- grey and persistant on the tree up to several years af- dominant with Keteleeria. -

Genetic Diversity and Population Structure of Pinus Kesiya Through Trans-Specific Amplification of Nuclear SSR Markers Kirti Chamling Rai*, H.S
International Journal of Agriculture, Environment and Biotechnology Citation: IJAEB: 11(5): 733-739, October 2018 DOI: 10.30954/0974-1712.10.2018.4 ©2018 New Delhi Publishers. All rights reserved GENETICS AND PLANT BREEDING Genetic Diversity and Population Structure of Pinus kesiya through Trans-specific Amplification of Nuclear SSR Markers Kirti Chamling Rai*, H.S. Ginwal and Romeet Saha Division of Genetics and Tree Propagation, Forest Research Institute, Dehradun, India *Corresponding author: [email protected] (ORCID ID: 0000-0001-5241-4334) Paper No. 737 Received: 20-07-2018 Accepted: 02-11-2018 ABSTRACT De novo primer development is cost intensive and time-consuming, therefore using primers developed for other species on the target species is a more preferred alternative. In the present study, a total of 47 primer pairs from P. taeda, P. merkusii, P. resinosa and P. densiflora were used for trans-specific amplification of P. kesiya. It was observed that only 5 (10.6%) primer pairs out of 47 transferred in P. kesiya which may be due to the phylogenetic distance of the target species from the source species. The expected heterozygosity (HE) ranged from 0.490 to 0.603 with a mean of 0.540 and the observed heterozygosity (HO) ranged from 0.044 to 0.819 with a mean of 0.342. The study has shown that the nuclear SSR markers can be utilized for estimating the genetic structure of P. kesiya populations. Results of the present work will go a long way in implementing proper strategies for the better management and conservation of P. kesiya forests and initiating tree improvement programmes in this species. -

Relationships Between Vegetation and Stomata, and Between Vegetation and Pollen Surface Soil in Yunnan, Southwest China
Article Ecology May 2013 Vol.58 No.15: 17751786 doi: 10.1007/s11434-012-5657-2 Relationships between vegetation and stomata, and between vegetation and pollen surface soil in Yunnan, Southwest China SHEN HuaDong1,2, LI ChunHai1*, WAN HeWen3, TONG GuoBang4, LIU JinSong5,6 & DAN Johnson7 1 State Key Laboratory of Lake Science and Environment, Nanjng Institute of Geography and Limnology, Chinese Academy of Sciences, Nanjing 210008, China; 2 University of Chinese Academy of Sciences, Beijing 100049, China; 3 College of Life Sciences, Anhui University, Hefei 230039, China; 4 Institute of Hydrogeology and Environmental Geology, Chinese Academy of Geological Sciences, Zhengding 050803, China; 5 Mathematics and Information Science College, Hebei Normal University, Shijiazhuang 050024, China; 6 The Key Laboratory of Calculation and Application of Hebei Province, Shijiazhuang 050024, China; 7 Environmental Science, Department of Geography, University of Lethbridge, Alberta T1K 3M4, Canada Received August 26, 2012; accepted November 30, 2012; published online February 21, 2013 Surface pollen and stomata of 61 samples collected in a study area ranging from tropical seasonal rainforest to oak forest (Quer- cus spinosa) in the Yulong Snow Mountain region in Yunnan, China, are used to distinguish vegetation communities. The results show that tropical seasonal rainforest (and mountain rainforest), south subtropical evergreen broad-leaved forest, and Quercus shrub are distinguished effectively from other vegetation types by analysis of surface pollen. The south subtropical evergreen broad-leaved forest, Pinus kesiya forest and evergreen broadleaf forest are distinguished effectively from other types of vegetation by pollen analysis. However, P. yunnanensis forest is not distinguished from other vegetation types, and P. -
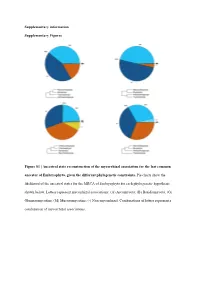
Ancestral State Reconstruction of the Mycorrhizal Association for the Last Common Ancestor of Embryophyta, Given the Different Phylogenetic Constraints
Supplementary information Supplementary Figures Figure S1 | Ancestral state reconstruction of the mycorrhizal association for the last common ancestor of Embryophyta, given the different phylogenetic constraints. Pie charts show the likelihood of the ancestral states for the MRCA of Embryophyta for each phylogenetic hypothesis shown below. Letters represent mycorrhizal associations: (A) Ascomycota; (B) Basidiomycota; (G) Glomeromycotina; (M) Mucoromycotina; (-) Non-mycorrhizal. Combinations of letters represent a combination of mycorrhizal associations. Austrocedrus chilensis Chamaecyparis obtusa Sequoiadendron giganteum Prumnopitys taxifolia Prumnopitys Prumnopitys montana Prumnopitys Prumnopitys ferruginea Prumnopitys Araucaria angustifolia Araucaria Dacrycarpus dacrydioides Dacrycarpus Taxus baccata Podocarpus oleifolius Podocarpus Afrocarpus falcatus Afrocarpus Ephedra fragilis Nymphaea alba Nymphaea Gnetum gnemon Abies alba Abies balsamea Austrobaileya scandens Austrobaileya Abies nordmanniana Thalictrum minus Thalictrum Abies homolepis Caltha palustris Caltha Abies magnifica ia repens Ranunculus Abies religiosa Ranunculus montanus Ranunculus Clematis vitalba Clematis Keteleeria davidiana Anemone patens Anemone Tsuga canadensis Vitis vinifera Vitis Tsuga mertensiana Saxifraga oppositifolia Saxifraga Larix decidua Hypericum maculatum Hypericum Larix gmelinii Phyllanthus calycinus Phyllanthus Larix kaempferi Hieronyma oblonga Hieronyma Pseudotsuga menziesii Salix reinii Salix Picea abies Salix polaris Salix Picea crassifolia Salix herbacea -

Carbon Stock Potential of Benguet Pine (Pinus Kesiya Royle Ex Gordon)
Ecosystems & Development Journal 7(2): 28–36 October 2017 ISSN 2012–3612 ABSTRACT The study evaluated the climate change mitigation potential of Benguet Pine (Pinus kesiya Royle ex Gordon) forests in a Carbon Stock Potential of mining site. It measured the amount of biomass, carbon and equivalent CO2 stored in trees, understorey and herbaceous (UH), necromass and litter (NL), coarse woody debris (CWD), Benguet Pine (Pinus kesiya roots and soil organic carbon (SOC) under three stand density types. It adopted a single stratum stratified sampling. A non– Royle ex Gordon) Stands destructive sampling was applied to all trees with diameter of 5 cm and above. Total harvesting of UH and NL samples were within a Mining Site in done in subplots inside the main sample plots. Composite soil Padcal, Benguet Province, samples were collected per soil depth (0–10 cm; 10–20 cm and 20–30 cm) and replicated per elevation type (high, mid and Philippines lower elevation). Stand types were classified using Google Earth image and GIS–generated map based on crown stocking density (CSD): high (HDS) = 2/3 CSD, medium (MDS) = > a* b 1/3 – < 2/3 CSD and low (LDS) = >1/3 CSD. Results showed Elenita L. Racelis , Diomedes A. Racelis , –1 b c a mean biomass density of 176.83 Mg ha with C content of Teodoro R. Villanueva , Leonardo M. Florece , –1 b b 169.74 Mg ha . The C content per stand type is as follows: Myrna G. Carandang , and Renato L. Lapitan HDS= 199.06 Mg ha–1; MDS = 188.91 Mg ha–1 and LDS = 123.03 Mg ha–1. -

Premier Phytoandrogen
IJBPAS, June, 2018, 7(6): 1121-1132 ISSN: 2277–4998 BENGUET PINE POLLEN (Pinus kesiya) AS NATURAL SOURCE OF PHYTOANDROGEN RAVELINA R. VELASCO1, DREXEL JAY M. DOLLENTE1, LEXTER R. NATIVIDAD2 AND TERESO A. ABELLA1 1College of Fisheries-Freshwater Aquaculture Center, Central Luzon State University, Science City of Muñoz, Nueva Ecija, Philippines 2 University Science High School, College of Education, Central Luzon State University, Science City of Muñoz, Nueva Ecija, Philippines *Corresponding author: Ravelina R. Velasco : E-mail: [email protected] Received 28th Feb. 2018; Revised 20th March 2018; Accepted 25th March 2018; Available online 1st June 2018 DOI: https://doi.org/10.31032/IJBPAS/2018/7.6.4472 ABSTRACT The result of the qualitative (colorimetric) phytochemical screening of pine tree indicated the presence of tannins (group of phenols), alkaloids (nitrogen containing compounds), steroids (groups of cholesterol, estrogen and testosterone), flavonoids (hydroxyl group), saponins (glycosidic/sugar compounds) and cardiac glycosides (secondary metabolites). All parts of pine tree (pine pollen, bark, twigs, roots, needles, branches) were tested positive for unsaturated steroids/sterols. The quantitative analysis of the pine pollen extract was performed using Gas chromatographic mass spectrometry. The compounds present in pine pollen were tricyclo (C9H12), pinene/careen/santolinatrienebicyclo (C10H16). These compounds are called terpenes, a hydrocarbon group. The testosterone content of pine pollen with an abundance of 1.25 x 106 was determined at 14 minutes. This testosterone content was confirmed when a standard 17- alpha methyltestosterone was also subjected to LCMS. Thin Layer Chromatography was also used to separate organic compounds. Under an ultra violet lamp, a change in the color of the yellow or brownish yellow spots in the developed chromatogram to red, violet, green or purple indicates the presence of phytochemicals. -

Plant Genetic Resources.Hwp
Korean J. Plant Res. 22(6) : 571~583 (2009) Review Plant Genetic Resources in Lam Dong province - Vietnam : Brief in medicine plants and wild orchids situation Nguyen Van Ket1* and Joon-Hyeong Cho2 1Agriculture and Forestry Faculty, The Dalat University, 01 Phu Dong Thien Vuong St. Lam Dong Province, Dalat City, Viet Nam 2Derparment of Plant Biotechnology, Dongguk University, Korea 26 Pil-dong 3-ga, Jung-gu, Seoul, 100-750, Korea Abstract - Lam Dong Province of Vietnam has an exceptional diversity of orchids and herbs. At least 920 herb species are widely mentioned in Vietnamese traditional medicinal literature and over 239 Lam Dong wild orchid species are used for attention under the Conservation Vietnamese wild orchids projects. In collaboration with Department of Plant Biotechnology, Dongguk University – South Korea working under supported ODA funds from Korea government (MIFAFF), we will consider how to collecting and preserving these plants in situ or conservating in vitro as a genetic resource. Key words - Vietnam, wild orchids, herbs Viet Nam geography and climate Topography and climate together with geology have brought in changes in the soil conditions; and their combined Viet Nam is approximately 331,688 km2 in area with the influence is well evident on the vegetation types and floristic perimeter of the country running along its international composition. Further and more complexity is manifested in boundaries is 4,639 km. Vietnam’s geographical location is the Vietnam’s flora as a result of southward extension of the in the tropical monsoon belt characterized by two seasons - flora of the South China and the Himalayas (Fisher, 1969).