Open Science and the Role of Publishers in Reproducible Research
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Book Self-Publishing Best Practices
Montana Tech Library Digital Commons @ Montana Tech Graduate Theses & Non-Theses Student Scholarship Fall 2019 Book Self-Publishing Best Practices Erica Jansma Follow this and additional works at: https://digitalcommons.mtech.edu/grad_rsch Part of the Communication Commons Book Self-Publishing Best Practices by Erica Jansma A project submitted in partial fulfillment of the requirements for the degree of M.S. Technical Communication Montana Tech 2019 ii Abstract I have taken a manuscript through the book publishing process to produce a camera-ready print book and e-book. This includes copyediting, designing layout templates, laying out the document in InDesign, and producing an index. My research is focused on the best practices and standards for publishing. Lessons learned from my research and experience include layout best practices, particularly linespacing and alignment guidelines, as well as the limitations and capabilities of InDesign, particularly its endnote functionality. Based on the results of this project, I can recommend self-publishers to understand the software and distribution platforms prior to publishing a book to ensure the required specifications are met to avoid complications later in the process. This document provides details on many of the software, distribution, and design options available for self-publishers to consider. Keywords: self-publishing, publishing, books, ebooks, book design, layout iii Dedication I dedicate this project to both of my grandmothers. I grew up watching you work hard, sacrifice, trust, and love with everything you have; it was beautiful; you are beautiful; and I hope I can model your example with a fraction of your grace and fruitfulness. Thank you for loving me so well. -

Reading Peer Review
EVE This Element describes for the first time the database of peer review reports at PLOS ONE, the largest scientific journal in ET AL. the world, to which the authors had unique access. Specifically, Reading Peer Review this Element presents the background contexts and histories of peer review, the data-handling sensitivities of this type PLOS ONE and Institutional of research, the typical properties of reports in the journal Change in Academia to which the authors had access, a taxonomy of the reports, and their sentiment arcs. This unique work thereby yields a compelling and unprecedented set of insights into the evolving state of peer review in the twenty-first century, at a crucial political moment for the transformation of science. It also, though, presents a study in radicalism and the ways in which Reading Peer Review Peer Reading PLOS’s vision for science can be said to have effected change in the ultra-conservative contemporary university. This title is also available as Open Access on Cambridge Core. Cambridge Elements in Publishing and Book Culture Series Editor: Samantha Rayner University College London Associate Editor: Leah Tether University of Bristol Publishing and Book Culture Academic Publishing Martin Paul Eve, Cameron Neylon, Daniel ISSN 2514-8524 (online) ISSN 2514-8516 (print) Paul O’Donnell, Samuel Moore, Robert Gadie, Downloaded from https://www.cambridge.org/core. IP address: 170.106.33.14, on 25 Sep 2021 at 19:03:04, subject to the Cambridge CoreVictoria terms of use, available Odeniyi at https://www.cambridge.org/core/terms and Shahina Parvin. https://doi.org/10.1017/9781108783521 Downloaded from https://www.cambridge.org/core. -

Gies for Increasing Citations
CORE Metadata, citation and similar papers at core.ac.uk Provided by International SERIES on Information Systems and Management in Creative eMedia (CreMedia) Maximized Research Impact: Effective Strate- gies for Increasing Citations Nader Ale Ebrahim*1, Hossein Gholizadeh2, Artur Lugmayr3 1Research Support Unit, Centre for Research Services, Institute of Research Management and Monitoring (IPPP), University of Malaya, Malaysia, Email: [email protected], *Corresponding author 2Department of Biomedical Engineering, Faculty of Engineering, University of Malaya, 50603 Kuala Lumpur, Malaysia 3VisLab., School of Media, Culture and Creative Arts, Curtin University, Perth, Australia Abstract The high competitive environment has forced higher education authori- ties to set their strategies to improve university ranking. Citations of published pa- pers are among the most widely used inputs to measure national and global uni- versity ranking (which accounts for 20% of QS, 30% of THE, and etc.). Therefore, from one hand, improving the citation impact of a search is one of the university manager’s strategies. On the other hand, the researchers are also looking for some helpful techniques to increase their citation record. This chapter by reviewing the relevant articles covers 48 different strategies for maximizing research impact and visibility. The results show that some features of article can help predict the num- ber of article views and citation counts. The findings presented in this chapter could be used by university authorities, authors, reviewers, and editors to maxim- ize the impact of articles in the scientific community. Keywords: University ranking, Improve citation, Citation frequency, Research impact, Open access, h-index Introduction The research output is an essential part of an institution’s measure and evaluation of research quality. -

Sci-Hub Provides Access to Nearly All Scholarly Literature
Sci-Hub provides access to nearly all scholarly literature A DOI-citable version of this manuscript is available at https://doi.org/10.7287/peerj.preprints.3100. This manuscript was automatically generated from greenelab/scihub-manuscript@51678a7 on October 12, 2017. Submit feedback on the manuscript at git.io/v7feh or on the analyses at git.io/v7fvJ. Authors • Daniel S. Himmelstein 0000-0002-3012-7446 · dhimmel · dhimmel Department of Systems Pharmacology and Translational Therapeutics, University of Pennsylvania · Funded by GBMF4552 • Ariel Rodriguez Romero 0000-0003-2290-4927 · arielsvn · arielswn Bidwise, Inc • Stephen Reid McLaughlin 0000-0002-9888-3168 · stevemclaugh · SteveMcLaugh School of Information, University of Texas at Austin • Bastian Greshake Tzovaras 0000-0002-9925-9623 · gedankenstuecke · gedankenstuecke Department of Applied Bioinformatics, Institute of Cell Biology and Neuroscience, Goethe University Frankfurt • Casey S. Greene 0000-0001-8713-9213 · cgreene · GreeneScientist Department of Systems Pharmacology and Translational Therapeutics, University of Pennsylvania · Funded by GBMF4552 PeerJ Preprints | https://doi.org/10.7287/peerj.preprints.3100v2 | CC BY 4.0 Open Access | rec: 12 Oct 2017, publ: 12 Oct 2017 Abstract The website Sci-Hub provides access to scholarly literature via full text PDF downloads. The site enables users to access articles that would otherwise be paywalled. Since its creation in 2011, Sci- Hub has grown rapidly in popularity. However, until now, the extent of Sci-Hub’s coverage was unclear. As of March 2017, we find that Sci-Hub’s database contains 68.9% of all 81.6 million scholarly articles, which rises to 85.2% for those published in toll access journals. -

Market Power in the Academic Publishing Industry
Market Power in the Academic Publishing Industry What is an Academic Journal? • A serial publication containing recent academic papers in a certain field. • The main method for communicating the results of recent research in the academic community. Why is Market Power important to think about? • Commercial academic journal publishers use market power to artificially inflate subscription prices. • This practice drains the resources of libraries, to the detriment of the public. How Does Academic Publishing Work? • Author writes paper and submits to journal. • Paper is evaluated by peer reviewers (other researchers in the field). • If accepted, the paper is published. • Libraries pay for subscriptions to the journal. The market does not serve the interests of the public • Universities are forced to “double-pay”. 1. The university funds research 2. The results of the research are given away for free to journal publishers 3. The university library must pay to get the research back in the form of journals Subscription Prices are Outrageous • The highest-priced journals are those in the fields of science, technology, and medicine (or STM fields). • Since 1985, the average price of a journal has risen more than 215 percent—four times the average rate of inflation. • This rise in prices, combined with the CA budget crisis, has caused UC Berkeley’s library to cancel many subscriptions, threatening the library’s reputation. A Comparison Why are prices so high? Commercial publishers use market power to charge inflated prices. Why do commercial publishers have market power? • They control the most prestigious, high- quality journals in many fields. • Demand is highly inelastic for high-quality journals. -

Google Scholar, Sci-Hub and Libgen: Could They Be Our New Partners?
Purdue University Purdue e-Pubs Proceedings of the IATUL Conferences 2017 IATUL Proceedings Google Scholar, Sci-Hub and LibGen: Could they be our New Partners? Louis Houle McGill University, [email protected] Louis Houle, "Google Scholar, Sci-Hub and LibGen: Could they be our New Partners?." Proceedings of the IATUL Conferences. Paper 3. https://docs.lib.purdue.edu/iatul/2017/partnership/3 This document has been made available through Purdue e-Pubs, a service of the Purdue University Libraries. Please contact [email protected] for additional information. GOOGLE SCHOLAR, SCI-HUB AND LIBGEN: COULD THEY BE OUR NEW PARTNERS? Louis Houle McGill University Canada [email protected] Abstract Since its debut I November 2004, librarians have raised several criticisms at Google Scholar (GS) such as its inconsistency of coverage and its currency and scope of coverage. It may have been true in the early years of Google Scholar but is this still through twelve years after? Is this sufficient to ignore it totally either in our information literacy programs or evaluate its value against the values of subscription-based abstracts and indexes? In this era of severe budget constraints that libraries are facing, can we imagine of substituting most or all of our subject databases with the free access of Google Scholar for discoverability? How much overlap between our databases and Google Scholar? How reliable is Google Scholar? How stable is its content over time? Open Access is getting to be the predominant form of getting access to peer reviewed articles. Many new non-traditional tools (institutional repositories, social media and peer to peer sites) are available out there to retrieve the full-text of peer reviewed articles. -
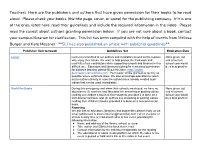
Teachers: Here Are the Publishers and Authors That Have Given Permission for Their Books to Be Read Aloud. Please Check Your Bo
Teachers: Here are the publishers and authors that have given permission for their books to be read aloud. Please check your books (the title page, cover, or spine) for the publishing company. If it is one of the ones listed here, read their guidelines and include the required information in the video. Please read the caveat about authors granting permission below. If you are not sure about a book, contact your campus librarian for clarification. This list has been compiled with the help of tweets from Melissa Burger and Kate Messner. **SLJ has also published an article with publisher guidelines** Publisher (link to tweet) Guidelines Set Expiration Date Lerner Lerner is committed to our authors and illustrators as well as the readers None given, but who enjoy their books. We want to help protect the hard work and end of current creativity of our contributors while supporting schools and libraries in this school year would difficult time. Educators and librarians looking for read-aloud permission be a best practice for distance learning, please fill out this form: https://rights- permissions.lernerbooks.com . Permission will be granted as quickly as possible where contracts allow. We also encourage educators to reach out to authors directly in case the authors have already created such videos that can be used immediately. MacMillan Books During this emergency and when their schools are closed, we have no None given, but objection to (1) teachers and librarians live streaming or posting videos end of current reading our children’s books to their students, provided it is done on a school year would noncommercial basis, and (2) authors live streaming or posting videos be a best practice reading their children’s books, provided it is done on a noncommercial basis. -

Exploratory Analysis of Publons Metrics and Their Relationship with Bibliometric and Altmetric Impact
Exploratory analysis of Publons metrics and their relationship with bibliometric and altmetric impact José Luis Ortega Institute for Advanced Social Studies (IESA-CSIC), Córdoba, Spain, [email protected] Abstract Purpose: This study aims to analyse the metrics provided by Publons about the scoring of publications and their relationship with impact measurements (bibliometric and altmetric indicators). Design/methodology/approach: In January 2018, 45,819 research articles were extracted from Publons, including all their metrics (scores, number of pre and post reviews, reviewers, etc.). Using the DOI identifier, other metrics from altmetric providers were gathered to compare the scores of those publications in Publons with their bibliometric and altmetric impact in PlumX, Altmetric.com and Crossref Event Data (CED). Findings: The results show that (1) there are important biases in the coverage of Publons according to disciplines and publishers; (2) metrics from Publons present several problems as research evaluation indicators; and (3) correlations between bibliometric and altmetric counts and the Publons metrics are very weak (r<.2) and not significant. Originality/value: This is the first study about the Publons metrics at article level and their relationship with other quantitative measures such as bibliometric and altmetric indicators. Keywords: Publons, Altmetrics, Bibliometrics, Peer-review 1. Introduction Traditionally, peer-review has been the most appropriate way to validate scientific advances. Since the first beginning of the scientific revolution, scientific theories and discoveries were discussed and agreed by the research community, as a way to confirm and accept new knowledge. This validation process has arrived until our days as a suitable tool for accepting the most relevant manuscripts to academic journals, allocating research funds or selecting and promoting scientific staff. -

Inter-Rater Reliability and Convergent Validity of F1000prime Peer Review
Accepted for publication in the Journal of the Association for Information Science and Technology Inter-rater reliability and convergent validity of F1000Prime peer review Lutz Bornmann Division for Science and Innovation Studies Administrative Headquarters of the Max Planck Society Hofgartenstr. 8, 80539 Munich, Germany. Email: [email protected] Abstract Peer review is the backbone of modern science. F1000Prime is a post-publication peer review system of the biomedical literature (papers from medical and biological journals). This study is concerned with the inter-rater reliability and convergent validity of the peer recommendations formulated in the F1000Prime peer review system. The study is based on around 100,000 papers with recommendations from Faculty members. Even if intersubjectivity plays a fundamental role in science, the analyses of the reliability of the F1000Prime peer review system show a rather low level of agreement between Faculty members. This result is in agreement with most other studies which have been published on the journal peer review system. Logistic regression models are used to investigate the convergent validity of the F1000Prime peer review system. As the results show, the proportion of highly cited papers among those selected by the Faculty members is significantly higher than expected. In addition, better recommendation scores are also connected with better performance of the papers. Keywords F1000; F1000Prime; Peer review; Highly cited papers; Adjusted predictions; Adjusted predictions at representative values 2 1 Introduction The first known cases of peer-review in science were undertaken in 1665 for the journal Philosophical Transactions of the Royal Society (Bornmann, 2011). Today, peer review is the backbone of science (Benda & Engels, 2011); without a functioning and generally accepted evaluation instrument, the significance of research could hardly be evaluated. -

Do You Speak Open Science? Resources and Tips to Learn the Language
Do You Speak Open Science? Resources and Tips to Learn the Language. Paola Masuzzo1, 2 - ORCID: 0000-0003-3699-1195, Lennart Martens1,2 - ORCID: 0000- 0003-4277-658X Author Affiliation 1 Medical Biotechnology Center, VIB, Ghent, Belgium 2 Department of Biochemistry, Ghent University, Ghent, Belgium Abstract The internet era, large-scale computing and storage resources, mobile devices, social media, and their high uptake among different groups of people, have all deeply changed the way knowledge is created, communicated, and further deployed. These advances have enabled a radical transformation of the practice of science, which is now more open, more global and collaborative, and closer to society than ever. Open science has therefore become an increasingly important topic. Moreover, as open science is actively pursued by several high-profile funders and institutions, it has fast become a crucial matter to all researchers. However, because this widespread interest in open science has emerged relatively recently, its definition and implementation are constantly shifting and evolving, sometimes leaving researchers in doubt about how to adopt open science, and which are the best practices to follow. This article therefore aims to be a field guide for scientists who want to perform science in the open, offering resources and tips to make open science happen in the four key areas of data, code, publications and peer-review. The Rationale for Open Science: Standing on the Shoulders of Giants One of the most widely used definitions of open science originates from Michael Nielsen [1]: “Open science is the idea that scientific knowledge of all kinds should be openly shared as early as is practical in the discovery process”. -

How Should Peer-Review Panels Behave? IZA DP No
IZA DP No. 7024 How Should Peer-Review Panels Behave? Daniel Sgroi Andrew J. Oswald November 2012 DISCUSSION PAPER SERIES Forschungsinstitut zur Zukunft der Arbeit Institute for the Study of Labor How Should Peer-Review Panels Behave? Daniel Sgroi University of Warwick Andrew J. Oswald University of Warwick and IZA Discussion Paper No. 7024 November 2012 IZA P.O. Box 7240 53072 Bonn Germany Phone: +49-228-3894-0 Fax: +49-228-3894-180 E-mail: [email protected] Any opinions expressed here are those of the author(s) and not those of IZA. Research published in this series may include views on policy, but the institute itself takes no institutional policy positions. The IZA research network is committed to the IZA Guiding Principles of Research Integrity. The Institute for the Study of Labor (IZA) in Bonn is a local and virtual international research center and a place of communication between science, politics and business. IZA is an independent nonprofit organization supported by Deutsche Post Foundation. The center is associated with the University of Bonn and offers a stimulating research environment through its international network, workshops and conferences, data service, project support, research visits and doctoral program. IZA engages in (i) original and internationally competitive research in all fields of labor economics, (ii) development of policy concepts, and (iii) dissemination of research results and concepts to the interested public. IZA Discussion Papers often represent preliminary work and are circulated to encourage discussion. Citation of such a paper should account for its provisional character. A revised version may be available directly from the author. -

Gigabyte: Publishing at the Speed of Research Scott C
EDITORIAL GigaByte: Publishing at the Speed of Research Scott C. Edmunds1,* and Laurie Goodman1 1 GigaScience, BGI Hong Kong Tech Co Ltd., 26F A Kings Wing Plaza, 1 On Kwan Street, Shek Mun, Sha Tin, NT, Hong Kong, China ABSTRACT Current practices in scientific publishing are unsuitable for rapidly changing fields and for presenting updatable data sets and software tools. In this regard, and as part of our continuing pursuit of pushing scientific publishing to match the needs of modern research, we are delighted to announce the launch of GigaByte, an online open-access, open data journal that aims to be a new way to publish research following the software paradigm: CODE, RELEASE, FORK, UPDATE and REPEAT. Following on the success of GigaScience in promoting data sharing and reproducibility of research, its new sister, GigaByte, aims to take this even further. With a focus on short articles, using a questionnaire-style review process, and combining that with the custom built publishing infrastructure from River Valley Technologies, we now have a cutting edge, XML-first publishing platform designed specifically to make the entire publication process easier, quicker, more interactive, and better suited to the speed needed to communicate modern research. Subjects Computer Sciences, Data Integration, Data Management FUTURE’S PAST In 2012, we launched GigaScience [1] as a new type of journal — one that provides standard scientific publishing linked directly to a database that hosts all the relevant data. Aiming to address Buckheit and Donoho’s 1995 complaint that “an article about computational results is advertising, not scholarship. The actual scholarship is the full software environment, code and data that produced the result.” [2], we were inspired to launch a new type of journal Submitted: 29 May 2020 that focussed on making sure the actual scholarship of all types of research was included Accepted: 01 June 2020 with the article narrative.