Levin and Miller 2005
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Phylogenetic Framework for Evolutionary Study of the Nightshades
Särkinen et al. BMC Evolutionary Biology 2013, 13:214 http://www.biomedcentral.com/1471-2148/13/214 RESEARCH ARTICLE Open Access A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree Tiina Särkinen1,2*, Lynn Bohs3, Richard G Olmstead4 and Sandra Knapp1 Abstract Background: The Solanaceae is a plant family of great economic importance. Despite a wealth of phylogenetic work on individual clades and a deep knowledge of particular cultivated species such as tomato and potato, a robust evolutionary framework with a dated molecular phylogeny for the family is still lacking. Here we investigate molecular divergence times for Solanaceae using a densely-sampled species-level phylogeny. We also review the fossil record of the family to derive robust calibration points, and estimate a chronogram using an uncorrelated relaxed molecular clock. Results: Our densely-sampled phylogeny shows strong support for all previously identified clades of Solanaceae and strongly supported relationships between the major clades, particularly within Solanum. The Tomato clade is shown to be sister to section Petota, and the Regmandra clade is the first branching member of the Potato clade. The minimum age estimates for major splits within the family provided here correspond well with results from previous studies, indicating splits between tomato & potato around 8 Million years ago (Ma) with a 95% highest posterior density (HPD) 7–10 Ma, Solanum & Capsicum c. 19 Ma (95% HPD 17–21), and Solanum & Nicotiana c. 24 Ma (95% HPD 23–26). Conclusions: Our large time-calibrated phylogeny provides a significant step towards completing a fully sampled species-level phylogeny for Solanaceae, and provides age estimates for the whole family. -

256. SOLANACEAE A. L. De Jussieu
256. SOLANACEAE A. L. de Jussieu SINOPSIS DE GÉNEROS Y TAXONES SUPRAGENÉRICOS DE ARGENTINA (Sistema de A. T. Hunziker, Adelaide, 1994) Subfamilia I. SOLANOIDEAE (15 gén.) Tribu I. Solaneae (9 gén.) Tribu III. Jaboroseae Miers (2 gén.) Solanum L. Jaborosa Juss. Aureliana Sendtn. Salpichroa Miers Capsicum L. Cyphomandra Sendtn. Dunalia Kunth Tribu IV. Lycieae Hunz. (2 gén.) Iochroma Benth. Lycianthes (Dunal) Hassler Lycium L. Physalis L. Grabowskia Schlecht. Vassobia Rusby Tribu V. Nicandreae Miers (1 gén.) Tribu II. Datureae Rchb. (1 gén.) Nicandra Adanson Datura L. Subfamilia II. CESTROIDEAE Schlecht. (17 gén.) Tribu VI. Cestreae G.Don (2 gén.) d. Subtribu Benthamiellinae Hunz. Cestrum L. (3 gén.) Sessea Ruiz et Pav. Benthamiella Speg. Combera Sandwith Tribu VII. Nicotianeae G. Don (9 gén.) Pantacantha Speg. a. Subtribu Nicotianinae (3 gén.) Tribu VIII. Salpiglossideae Benth. (2 gén.) Nicotiana L. Salpiglossis Ruiz et Pav. Fabiana Ruiz et Pav. Reyesia Clos Petunia Juss. Tribu IX. Francisceae G. Don (1 gén.) b. Subtribu Nierembergiinae Hunz. et Andr. Cocucci (2 gén.) Brunfelsia L. Nierembergia Ruiz et Pav. Tribu X. Schwenckieae Hunz. (2 gén.) Bouchetia Dunal Schwenckia L. c. Subtribu Leptoglossinae Hunz. Melananthus Walp. (1 gén.) Tribu XI. Schizantheae Miers (1 gén.) Leptoglossis Benth. Schizanthus Ruiz et Pav. Solanoideae: 5 tribus; 15 géneros Cestroideae: 6 tribus; 17 géneros TOTAL: 2 subfamilia; 11 tribus; 32 géneros 256. SOLANACEAE Tribu IV. LYCIEAE Hunz., parte B 2. Grabowskia Schlecht. 1, 2 D. F. L. Schlechtendahl, Linnaea 7: 71. 1832; etimol.: en honor del farmacéutico H. Grabowski, coautor de una flora regional del este europeo. Pukanthus Rafinesque, Sylva Tellur. Mant. Synopt.: 53. -

Seeds and Plants
r. i. -20. U. S. DEPARTMENT OF AGRICULTURE. SECTION OF SKKI) AND PLANT INTRODUCTION. INVENTORY NO. 8. SEEDS AND PLANTS, IMI'ORTED FOR DISTRIBUTION IN COOPERATION WITH THE AGRICULTURAL EXPERIMENT STATIONS. NUMBE11S 3401-4350. 10183—00 1 INVENTORY OF FOREIGN SEEDS AND PLANTS. INTRODUCTORY STATEMENT. This inventory or catalogue of seeds and plants includes a number of exceptionally valuable items collected by the Agricultural Explorers of the Section of Seed and Plant Introduction. There is an interest- ing and valuable series of economic plants of the most varied uses procured by the Hon. Harbour Lathrop, of Chicago, assisted by Mr. David G. Fairchild. Mr. W. T. Swingle has continued his work in Algeria, Sicily, and Turkey, and this list contains many of his impor- tations. There are also a number of donations from various sources, and a few seeds purchased directly from the growers. The following importations represent perhaps the most valuable of the many interesting novelties here described: Mr. Swingle's col- lection of improved varieties of the date palm, procured in Algeria; a collection of spineless cacti from the Argentine Republic secured by Messrs. Lathrop and Fairchild, which may become valuable forage plants in the arid Southwest; genge clover, a leguminous forage crop and green manure which is grown in the rice fields of Japan as a winter soil cover and fertilizer; a collection of broad beans from England, this vegetable being practically unknown in the United States, although extensively used in Europe and on the Continent; a new seedless raisin grape from Italy for the raisin growers of California and Arizona; a little sample of wheat from Peru, donated by Dr. -

A Molecular Phylogeny of the Solanaceae
TAXON 57 (4) • November 2008: 1159–1181 Olmstead & al. • Molecular phylogeny of Solanaceae MOLECULAR PHYLOGENETICS A molecular phylogeny of the Solanaceae Richard G. Olmstead1*, Lynn Bohs2, Hala Abdel Migid1,3, Eugenio Santiago-Valentin1,4, Vicente F. Garcia1,5 & Sarah M. Collier1,6 1 Department of Biology, University of Washington, Seattle, Washington 98195, U.S.A. *olmstead@ u.washington.edu (author for correspondence) 2 Department of Biology, University of Utah, Salt Lake City, Utah 84112, U.S.A. 3 Present address: Botany Department, Faculty of Science, Mansoura University, Mansoura, Egypt 4 Present address: Jardin Botanico de Puerto Rico, Universidad de Puerto Rico, Apartado Postal 364984, San Juan 00936, Puerto Rico 5 Present address: Department of Integrative Biology, 3060 Valley Life Sciences Building, University of California, Berkeley, California 94720, U.S.A. 6 Present address: Department of Plant Breeding and Genetics, Cornell University, Ithaca, New York 14853, U.S.A. A phylogeny of Solanaceae is presented based on the chloroplast DNA regions ndhF and trnLF. With 89 genera and 190 species included, this represents a nearly comprehensive genus-level sampling and provides a framework phylogeny for the entire family that helps integrate many previously-published phylogenetic studies within So- lanaceae. The four genera comprising the family Goetzeaceae and the monotypic families Duckeodendraceae, Nolanaceae, and Sclerophylaceae, often recognized in traditional classifications, are shown to be included in Solanaceae. The current results corroborate previous studies that identify a monophyletic subfamily Solanoideae and the more inclusive “x = 12” clade, which includes Nicotiana and the Australian tribe Anthocercideae. These results also provide greater resolution among lineages within Solanoideae, confirming Jaltomata as sister to Solanum and identifying a clade comprised primarily of tribes Capsiceae (Capsicum and Lycianthes) and Physaleae. -

Phylogenetics of the Caprifolieae and Lonicera (Dipsacales)
Systematic Botany (2008), 33(4): pp. 776–783 © Copyright 2008 by the American Society of Plant Taxonomists Phylogenetics of the Caprifolieae and Lonicera (Dipsacales) Based on Nuclear and Chloroplast DNA Sequences Nina Theis,1,3,4 Michael J. Donoghue,2 and Jianhua Li1,4 1Arnold Arboretum of Harvard University, 22 Divinity Ave, Cambridge, Massachusetts 02138 U.S.A. 2Department of Ecology and Evolutionary Biology, Yale University, P.O. Box 208106, New Haven, Conneticut 06520-8106 U.S.A. 3Current Address: Plant Soil, and Insect Sciences, University of Massachusetts at Amherst, Amherst, Massachusetts 01003 U.S.A. 4Authors for correspondence ([email protected]; [email protected]) Communicating Editor: Lena Struwe Abstract—Recent phylogenetic analyses of the Dipsacales strongly support a Caprifolieae clade within Caprifoliaceae including Leycesteria, Triosteum, Symphoricarpos, and Lonicera. Relationships within Caprifolieae, however, remain quite uncertain, and the monophyly of Lonicera, the most species-rich of the traditional genera, and its subdivisions, need to be evaluated. In this study we used sequences of the ITS region of nuclear ribosomal DNA and five chloroplast non-coding regions (rpoB–trnC spacer, atpB–rbcL spacer, trnS–trnG spacer, petN–psbM spacer, and psbM–trnD spacer) to address these problems. Our results indicate that Heptacodium is sister to Caprifolieae, Triosteum is sister to the remaining genera within the tribe, and Leycesteria and Symphoricarpos form a clade that is sister to a monophyletic Lonicera. Within Lonicera, the major split is between subgenus Caprifolium and subgenus Lonicera. Within subgenus Lonicera, sections Coeloxylosteum, Isoxylosteum, and Nintooa are nested within the paraphyletic section Isika. Section Nintooa may also be non-monophyletic. -

Kohn JR (2004) Historical Inferences from the Self-Incompatibility Locus
Chapter 5 What Genealogies of S-alleles Tell Us J.R. Kohn Abstract Drawing on examples from S-RNase-based self-incompatibility (SI), S- locus genealogies are used to infer the demographic history of lineages, the history of mating-system transitions in entire plant families and aspects of the evolution of the S-locus itself. Two lineages of Solanaceae suffered severe restrictions of S-locus diversity evident after millions of years. Broadly shared ancestral S-locus polymorphism is evidence that loss of this form of incompatibility was irreversible in the Solanaceae. Frequent and irreversible loss implies incompatibility is either declining in frequency through time, or that it confers an increased diversification rate relative to self-compatibility (SC). Differences in diversification rate among self-incompatible and self-compatible lineages likely cause the failure of current phylogenetic methods to correctly reconstruct the history of SI. Genealogies also show that origination of new S-RNases rarely occurs within the lifetimes of species. Surprisingly, genealogies of F-box genes purported to provide pollen specificity often do not correspond to those of their cognate S-RNases, indicating we have much to learn about how this system works and evolves. Abbreviations cpDNA DNA from plant plastids GSI Gametophytic SI mya Million years ago SC Self-compatibility SCR/SP11 S-locus cysteine-rich protein (the pollen S-determinant in Brassica) J.R. Kohn Section of Ecology, Behavior and Evolution, Division of Biological Sciences, University of California San Diego, 9500 Gilman Drive, La Jolla CA, 92093–0116 USA, e-mail: [email protected] V.E. Franklin-Tong (ed.) Self-Incompatibility in Flowering Plants – Evolution, Diversity, 103 and Mechanisms. -
B32-W6-Lecture Copy.Pptx
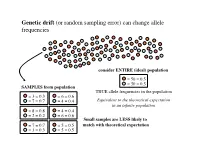
Genetic drift (or random sampling error) can change allele frequencies! consider ENTIRE (ideal) population! = 50 = 0.5! = 50 = 0.5! SAMPLES from population! TRUE allele frequencies in the population! = 3 = 0.3! = 6 = 0.6! = 7 = 0.7! = 4 = 0.4! Equivalent to the theoretical expectation! in an infinite population! = 8 = 0.8! = 4 = 0.4! = 2 = 0.2! = 6 = 0.6! Small samples are LESS likely to = 7 = 0.7! = 5 = 0.5! match with theoretical expectation! = 3 = 0.3! = 5 = 0.5! Random walks... describe fluctuations in the frequencies of alleles that are neutral with respect to fitness! demes ! Over generations, random sampling error occurs...! During each generation, the large number of zygotes produced is reduced (via mortality that is random with respect to fitness) to some number of breeding individuals! – allele frequencies change (drift) –! What happens to the frequencies of p and q over the long term...?! Alleles drift to fixation in each deme. ! Which allele gets fixed...?! The theory of genetic drift laid out by Sewell Wright during the 1930s and 1940s. Taken up by Motoo Kimura in the 1950s.! Natural selection & genetic drift are the 2 most important causes of evolutionary change in populations. ! The relative importance of each remains controversial.! Genetic drift as a null hypothesis... All alleles @ all loci are subject to genetic drift. The same cannot be said for natural selection.! Evolution by genetic drift...! (1)! Allele frequencies fluctuate at random within a population & eventually one (or another) allele will be fixed.! (2)! The magnitude of change is inversely related to the size of the sample; drift is stronger in smaller samples.! (3)! Drift is NOT repeatable. -

Download Manuscript Manuscript Datphypaper V23 Cleanversion.Docx
Manuscript Clean Version Click here to download Manuscript Manuscript_DatPhyPaper_v23_CleanVersion.docx 1 Phylogenetics of Datureae (Solanaceae), including description of the new genus Trompettia 2 and re-circumscription of the tribe 3 4 Julia Dupin1* and Stacey D. Smith1 5 6 1 Department of Ecology and Evolutionary Biology, University of Colorado Boulder, 1800 7 Colorado Ave, Boulder, CO 80309, USA. 8 * Correspondence author: [email protected] 9 10 Running head: Phylogenetics of Datureae (Solanaceae) 11 12 Abstract 13 14 Datureae G. Don is a tribe in the Solanaceae known for its charismatic large-flowered 15 species (jimsonweeds and angel trumpets). The monophyly of the tribe is well established, but the 16 recent finding that a species previously described in Iochroma (also Solanaceae) belongs in 17 Datureae calls for a reassessment of the tribe’s circumscription. Here we estimated the phylogeny of 18 Datureae, including of all of its 18 species, using three nuclear regions, and incorporated fossil 19 information to estimate divergence times. Based on this phylogeny, we reconstructed the evolution 20 of key aspects of reproductive morphology and life history to identify diagnostic features. Our 21 molecular phylogenetic analyses suggest that the diversification of Datureae began roughly ca. 35 22 Ma, around the beginning of the Andean uplift. Within the tribe, Datura and Brugmansia are 23 monophyletic sister taxa and the misplaced species of Iochroma is sister to the remaining species. 24 Based on our morphological analysis, we describe the latter as a new monotypic genus Trompettia. 25 Ancestral state reconstructions identify diagnostic features for each of the three genera and show a 26 large suite of changes along the Datura branch, including the evolution of erect flowers, capsular 27 fruit and annual life history. -

Flora of Australia, Volume 29, Solanaceae
FLORA OF AUSTRALIA Volume 29 Solanaceae This volume was published before the Commonwealth Government moved to Creative Commons Licensing. © Commonwealth of Australia 1982. This work is copyright. You may download, display, print and reproduce this material in unaltered form only (retaining this notice) for your personal, non-commercial use or use within your organisation. Apart from any use as permitted under the Copyright Act 1968, no part may be reproduced or distributed by any process or stored in any retrieval system or data base without prior written permission from the copyright holder. Requests and inquiries concerning reproduction and rights should be addressed to: [email protected] FLORA OF AUSTRALIA In this volume all 206 species of the family Solanaceae known to be indigenous or naturalised in Australia are described. The family includes important toxic plants, weeds and drug plants. The family Solanaceae in Australia contains 140 indigenous species such as boxthorn, wild tobacco, wild tomato, Pituri and tailflower. The 66 naturalised members include nightshade, tomato, thornapple, petunia, henbane, capsicum and Cape Gooseberry. There are keys for the identification of all genera and species. References are given for accepted names and synonyms. Maps are provided showing the distribution of nearly all species. Many are illustrated by line drawings or colour plates. Notes on habitat, variation and relationships are included. The volume is based on the most recent taxonomic research on the Solanaceae in Australia. Cover: Solanum semiarmatum F . Muell. Painting by Margaret Stones. Reproduced by courtesy of David Symon. Contents of volumes in the Flora of Australia, the faiimilies arranged according to the system of A.J. -

Bayesian Estimation of the Global Biogeographical History of The
Journal of Biogeography (J. Biogeogr.) (2016) ORIGINAL Bayesian estimation of the global ARTICLE biogeographical history of the Solanaceae Julia Dupin1*, Nicholas J. Matzke2, Tiina S€arkinen3, Sandra Knapp4, Richard G. Olmstead5, Lynn Bohs6 and Stacey D. Smith1 1Department of Ecology and Evolutionary ABSTRACT Biology, University of Colorado Boulder, Aim The tomato family Solanaceae is distributed on all major continents Boulder, CO 80309, USA, 2Division of except Antarctica and has its centre of diversity in South America. Its world- Ecology, Evolution and Genetics, Research School of Biology, The Australian National wide distribution suggests multiple long-distance dispersals within and between University, ACT 2601 Canberra, Australia, the New and Old Worlds. Here, we apply maximum likelihood (ML) methods 3Royal Botanic Garden Edinburgh, Edinburgh and newly developed biogeographical stochastic mapping (BSM) to infer the EH3 5LR, UK, 4Department of Life Sciences, ancestral range of the family and to estimate the frequency of dispersal and Natural History Museum, London SW7 5BD, vicariance events resulting in its present-day distribution. 5 UK, Department of Biology and Burke Location Worldwide. Museum, University of Washington, Seattle, Washington 98195, USA, 6Department of Methods Building on a recently inferred megaphylogeny of Solanaceae, we Biology, University of Utah, Salt Lake City, conducted ML model fitting of a range of biogeographical models with the UT 84112, USA program ‘BioGeoBEARS’. We used the parameters from the best fitting model to estimate ancestral range probabilities and conduct stochastic mapping, from which we estimated the number and type of biogeographical events. Results Our best model supported South America as the ancestral area for the Solanaceae and its major clades. -

Phylogeny of Nolana (Nolaneae, Solanoideae, Solanaceae) As Inferred from Granule-Bound Starch Synthase I (GBSSI) Sequences
Dillon & al. • Phylogeny of Nolana TAXON 56 (4) • November 2007: 1000–1011 Phylogeny of Nolana (Nolaneae, Solanoideae, Solanaceae) as inferred from granule-bound starch synthase I (GBSSI) sequences Michael O. Dillon1, Tieyao Tu2, Akiko Soejima3, Tingshuang Yi4, Zelong Nie2, Alan Tye5 & Jun Wen2,6 1 Botany Department, The Field Museum, 1400 South Lake Shore Drive, Chicago, Illinois 60605, U.S.A. [email protected] (author for correspondence) 2 Key Laboratory of Biodiversity and Biogeography, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650204, P. R. China; Graduate School of Chinese Academy of Sciences, Beijing 100039, P. R. China 3 School of Science, Osaka Prefecture University, Sakai, Osaka 599-8531, Japan 4 Key Laboratory of Biodiversity and Biogeography, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650204, P. R. China 5 Department of Botany, Charles Darwin Research Station, Isla Santa Cruz, Galápagos, Ecuador 6 Department of Botany, MRC-166 Smithsonian Institution, P.O. Box 37012, Washington, D.C. 20013-7012, U.S.A. The phylogenetic relationships of Nolana (Nolaneae, Solanaceae) were constructed using partial sequences (ca. 891 bp) of the granule-bound starch synthase I (GBSSI) or the waxy gene. Nolana, with 89 species, is primarily distributed in coastal Chile (49 spp.) and Peru (43 spp.), and of these, four species are recorded in Peru and Chile, and another from the Galápagos Islands, Ecuador. Our phylogenetic analysis, utilizing a sampling of 63 of the 89 species, supports the monophyly of Nolana and recovered three clades with 95%–100% bootstrap support. Nolana sessiliflora is the sister taxon to the remainder of the genus. -

Biology of Invasive Plants 2. Lycium Ferocissimum Miers
Invasive Plant Science and Biology of Invasive Plants 2. Lycium Management ferocissimum Miers www.cambridge.org/inp Michael R. Noble1, Robin J. Adair2 and Kylie B. Ireland3 1Program Coordinator (Invasive Species), Biosecurity Operations Branch, Department of Primary Industries, Parks, Water and Environment, Devonport, Tasmania, Australia; 2Director, Australis Biological, Bittern, Victoria, Australia Biology of Invasive Plants and 3Postdoctoral Fellow, Commonwealth Scientific and Industrial Research Organisation (CSIRO) Health and Biosecurity, Black Mountain Science & Innovation Park, Acton, Australian Capital Territory, Australia; current: Cite this article: Noble MR, Adair RJ, and Adjunct research fellow, Centre for Crop and Disease Management (CCDM), Curtin University, Bentley, Western Ireland KB (2021) Biology of Invasive Plants 2. Lycium ferocissimum Miers. Invasive Plant Sci. Australia, Australia; Plant pathologist, Department of Primary Industries and Development, South Perth, Manag 14:41–56. doi: 10.1017/inp.2021.13 Western Australia Received: 31 January 2021 Accepted: 4 March 2021 Scientific Classification Series Editors: Domain: Eukaryota Darren J. Kriticos, CSIRO Ecosystem Sciences & Kingdom: Plantae David R. Clements, Trinity Western University Phylum: Spermatophyta Keywords: Subphylum: Angiospermae African boxthorn; invasive plant Class: Dicotyledonae Order: Solanales Author for correspondence: Family: Solanaceae Michael R. Noble, Program Coordinator Subfamily: Solanoideae (Invasive Species), Biosecurity Operations Branch,