Regulation of Leukocytes by Tspanc8 Tetraspanins And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Comparing Spatial Expression Dynamics of Bovine
Comparing spatial expression dynamics of bovine blastocyst under three different procedures : in-vivo, in-vitro derived, Title and somatic cell nuclear transfer embryos Nagatomo, Hiroaki; Akizawa, Hiroki; Sada, Ayari; Kishi, Yasunori; Yamanaka, Ken-ichi; Takuma, Tetsuya; Sasaki, Author(s) Keisuke; Yamauchi, Nobuhiko; Yanagawa, Yojiro; Nagano, Masashi; Kono, Tomohiro; Takahashi, Masashi; Kawahara, Manabu Citation Japanese Journal of Veterinary Research, 63(4), 159-171 Issue Date 2015-11 DOI 10.14943/jjvr.63.4.159 Doc URL http://hdl.handle.net/2115/60304 Type bulletin (article) Additional Information There are other files related to this item in HUSCAP. Check the above URL. File Information JJVR63-4 p.159-171 Suppl. Table 3.pdf (Supplemental Table 3) Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP Supplemental table 3. Genes that were differentially expressed in the ICM relative to the TE in in SCNT blastocyst (SCNT list). Gene Symbol Probe Set ID Regulation Fold change ([ICM] / [TE]) Gene Title EEF1A1 AFFX-Bt-ef1a-3_at UP 1.2092365 eukaryotic translation elongation factor 1 alpha 1 IGFBP3 Bt.422.1.S2_at UP 2.5323892 insulin-like growth factor binding protein 3 IGFBP3 Bt.422.1.S1_at UP 3.7850845 insulin-like growth factor binding protein 3 SULT1A1 Bt.3537.1.S1_at UP 2.7092714 sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 SPP1 Bt.2632.1.S1_at UP 5.6928325 secreted phosphoprotein 1 SCARB1 Bt.4520.1.S1_at UP 3.106944 scavenger receptor class B, member 1 TSPO Bt.3988.1.S1_at -

Downloaded 18 July 2014 with a 1% False Discovery Rate (FDR)
UC Berkeley UC Berkeley Electronic Theses and Dissertations Title Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer Permalink https://escholarship.org/uc/item/0t47b9ws Author Spiciarich, David Publication Date 2017 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer by David Spiciarich A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Chemistry in the Graduate Division of the University of California, Berkeley Committee in charge: Professor Carolyn R. Bertozzi, Co-Chair Professor David E. Wemmer, Co-Chair Professor Matthew B. Francis Professor Amy E. Herr Fall 2017 Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer © 2017 by David Spiciarich Abstract Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer by David Spiciarich Doctor of Philosophy in Chemistry University of California, Berkeley Professor Carolyn R. Bertozzi, Co-Chair Professor David E. Wemmer, Co-Chair Changes in glycosylation have long been appreciated to be part of the cancer phenotype; sialylated glycans are found at elevated levels on many types of cancer and have been implicated in disease progression. However, the specific glycoproteins that contribute to cell surface sialylation are not well characterized, specifically in bona fide human cancer. Metabolic and bioorthogonal labeling methods have previously enabled enrichment and identification of sialoglycoproteins from cultured cells and model organisms. The goal of this work was to develop technologies that can be used for detecting changes in glycoproteins in clinical models of human cancer. -

Impact of Porcine Arterivirus, Influenza B, and Their Coinfection on Antiviral Response in the Porcine Lung
pathogens Article Impact of Porcine Arterivirus, Influenza B, and Their Coinfection on Antiviral Response in the Porcine Lung Damarius S. Fleming 1,2, Laura C. Miller 2,*, Yun Tian 3, Yonghai Li 4, Wenjun Ma 4,5 and Yongming Sang 3,* Article 1 Oak Ridge InstituteImpact for Science of Porcine and Education Arterivirus, Oakridge, Oak Ridge Influenza Associated Universities,B, and Their Oak Ridge, TNCoinfection 37830, USA; damarius.fl[email protected] on Antiviral Response in the Porcine 2 Virus and Prion Research Unit, National Animal Disease Center, USDA, Agricultural Research Service, Ames, IA 50161,Lung USA 3 Department of Agricultural and Environmental Sciences, Tennessee State University, Damarius S. Fleming 1,2, Laura C. Miller 2,*, Yun Tian 3, Yonghai Li 4, Wenjun Ma 4,5 and Nashville, TN 37209, USA; [email protected] Yongming Sang 3,* 4 Department of Diagnostic Medicine and Pathobiology, Kansas State University, Manhattan, KS 66506, USA; 1 [email protected] Oak (Y.L.); Ridge Institute [email protected] for Science and Education (W.M.) Oakridge, Oak Ridge Associated Universities, Oak Ridge, 5 TN 37830, USA; [email protected] Department of2 VeterinaryVirus and Prion Pathobiology Research Unit, and National Department Animal Disease of Molecular Center, USDA Microbiology, Agricultural & Research Immunology, Service, University of Missouri,Ames, IA Columbia, 50161, USA MO 65211, USA * Correspondence:3 Department [email protected] of Agricultural and (L.C.M.); Environmental [email protected] Sciences, Tennessee (Y.S.) State -

Tetraspanins in the Regulation of Mast Cell Function
Medical Microbiology and Immunology (2020) 209:531–543 https://doi.org/10.1007/s00430-020-00679-x REVIEW Tetraspanins in the regulation of mast cell function Zane Orinska1 · Philipp M. Hagemann1 · Ivana Halova2 · Petr Draber2 Received: 17 February 2020 / Accepted: 6 May 2020 / Published online: 7 June 2020 © The Author(s) 2020 Abstract Mast cells (MCs) are long-living immune cells highly specialized in the storage and release of diferent biologically active compounds and are involved in the regulation of innate and adaptive immunity. MC degranulation and replacement of MC granules are accompanied by active membrane remodelling. Tetraspanins represent an evolutionary conserved family of transmembrane proteins. By interacting with lipids and other membrane and intracellular proteins, they are involved in organisation of membrane protein complexes and act as “molecular facilitators” connecting extracellular and cytoplasmic signaling elements. MCs express diferent tetraspanins and MC degranulation is accompanied by changes in membrane organisation. Therefore, tetraspanins are very likely involved in the regulation of MC exocytosis and membrane reorganisa- tion after degranulation. Antiviral response and production of exosomes are further aspects of MC function characterized by dynamic changes of membrane organization. In this review, we pay a particular attention to tetraspanin gene expression in diferent human and murine MC populations, discuss tetraspanin involvement in regulation of key MC signaling com- plexes, and analyze the potential contribution of tetraspanins to MC antiviral response and exosome production. In-depth knowledge of tetraspanin-mediated molecular mechanisms involved in diferent aspects of the regulation of MC response will be benefcial for patients with allergies, characterized by overwhelming MC reactions. -

Regulation of ADAM10 by the Tspanc8 Family of Tetraspanins and Their Therapeutic Potential
International Journal of Molecular Sciences Review Regulation of ADAM10 by the TspanC8 Family of Tetraspanins and Their Therapeutic Potential Neale Harrison 1, Chek Ziu Koo 1,2 and Michael G. Tomlinson 1,2,* 1 School of Biosciences, University of Birmingham, Birmingham B15 2TT, UK; [email protected] (N.H.); [email protected] (C.Z.K.) 2 Centre of Membrane Proteins and Receptors (COMPARE), Universities of Birmingham and Nottingham, Midlands, UK * Correspondence: [email protected]; Tel.: +44-(0)121-414-2507 Abstract: The ubiquitously expressed transmembrane protein a disintegrin and metalloproteinase 10 (ADAM10) functions as a “molecular scissor”, by cleaving the extracellular regions from its membrane protein substrates in a process termed ectodomain shedding. ADAM10 is known to have over 100 substrates including Notch, amyloid precursor protein, cadherins, and growth factors, and is important in health and implicated in diseases such as cancer and Alzheimer’s. The tetraspanins are a superfamily of membrane proteins that interact with specific partner proteins to regulate their intracellular trafficking, lateral mobility, and clustering at the cell surface. We and others have shown that ADAM10 interacts with a subgroup of six tetraspanins, termed the TspanC8 subgroup, which are closely related by protein sequence and comprise Tspan5, Tspan10, Tspan14, Tspan15, Tspan17, and Tspan33. Recent evidence suggests that different TspanC8/ADAM10 complexes have distinct substrates and that ADAM10 should not be regarded as a single scissor, but as six different TspanC8/ADAM10 scissor complexes. This review discusses the published evidence for this “six Citation: Harrison, N.; Koo, C.Z.; scissor” hypothesis and the therapeutic potential this offers. -

Identification of Genomic Targets of Krüppel-Like Factor 9 in Mouse Hippocampal
Identification of Genomic Targets of Krüppel-like Factor 9 in Mouse Hippocampal Neurons: Evidence for a role in modulating peripheral circadian clocks by Joseph R. Knoedler A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Neuroscience) in the University of Michigan 2016 Doctoral Committee: Professor Robert J. Denver, Chair Professor Daniel Goldman Professor Diane Robins Professor Audrey Seasholtz Associate Professor Bing Ye ©Joseph R. Knoedler All Rights Reserved 2016 To my parents, who never once questioned my decision to become the other kind of doctor, And to Lucy, who has pushed me to be a better person from day one. ii Acknowledgements I have a huge number of people to thank for having made it to this point, so in no particular order: -I would like to thank my adviser, Dr. Robert J. Denver, for his guidance, encouragement, and patience over the last seven years; his mentorship has been indispensable for my growth as a scientist -I would also like to thank my committee members, Drs. Audrey Seasholtz, Dan Goldman, Diane Robins and Bing Ye, for their constructive feedback and their willingness to meet in a frequently cold, windowless room across campus from where they work -I am hugely indebted to Pia Bagamasbad and Yasuhiro Kyono for teaching me almost everything I know about molecular biology and bioinformatics, and to Arasakumar Subramani for his tireless work during the home stretch to my dissertation -I am grateful for the Neuroscience Program leadership and staff, in particular -

The Emerging Role of Tetraspanins in the Proteolytic Processing of the Amyloid Precursor Protein
MINI REVIEW published: 21 December 2016 doi: 10.3389/fnmol.2016.00149 The Emerging Role of Tetraspanins in the Proteolytic Processing of the Amyloid Precursor Protein Lisa Seipold and Paul Saftig * Institut für Biochemie, Christian-Albrechts-Universität zu Kiel (CAU), Kiel, Germany Tetraspanins are a family of ubiquitously expressed and conserved proteins, which are characterized by four transmembrane domains and the formation of a short and a large extracellular loop (LEL). Through interaction with other tetraspanins and transmembrane proteins such as growth factors, receptors and integrins, tetraspanins build a wide ranging and membrane spanning protein network. Such tetraspanin-enriched microdomains (TEMs) contribute to the formation and stability of functional signaling complexes involved in cell activation, adhesion, motility, differentiation, and malignancy. There is increasing evidence showing that the tetraspanins also regulate the proteolysis of the amyloid precursor protein (APP) by physically interacting with the APP secretases. CD9, CD63, CD81, Tspan12, Tspan15 are among the tetraspanins involved in the intracellular transport and in the stabilization of the gamma secretase complex or ADAM10 as the major APP alpha secretase. They also directly regulate, most likely in concert with other tetraspanins, the proteolytic function of these membrane embedded enzymes. Despite the knowledge about the interaction of tetraspanins with the Edited by: secretases not much is known about their physiological role, their importance in Thomas Deller, Goethe-University, Germany Alzheimer’s Disease and their exact mode of action. This review aims to summarize the Reviewed by: current knowledge and open questions regarding the biology of tetraspanins and the Monica DiLuca, understanding how these proteins interact with APP processing pathways. -

Gene List HTG Edgeseq Oncology Biomarker Panel
Gene List HTG EdgeSeq Oncology Biomarker Panel For Research Use Only. Not for use in diagnostic procedures. A2M ADRA2B APH1B BAG1 BRCA2 CARM1 CCNH CDC25A CHI3L1 COX7B CXCL16 DESI1 ABCA2 ADRA2C APOC2 BAG2 BRIP1 CASP1 CCNO CDC25B CHI3L2 CP CXCL2 DFFA ABCA3 AFF1 APOC4 BAG3 BTC CASP10 CCNT1 CDC25C CHMP4B CPT1A CXCL3 DHCR24 ABCA4 AGER APOL3 BAG4 BTG1 CASP12 CCR1 CDC34 CHPT1 CPT1B CXCL5 DHH ABCA5 AGFG1 APP BAG5 BTG2 CASP14 CCR10 CDC42 CHRNA1 CPT1C CXCL6 DHX58 ABCA9 AGGF1 APPBP2 BAI1 BTG3 CASP2 CCR2 CDC42BPA CHRNB1 CPT2 CXCL8 DIABLO ABCB11 AGT AQP1 BAIAP3 BTK CASP3 CCR3 CDC6 CHSY1 CRADD CXCL9 DIAPH3 ABCB4 AHNAK AQP2 BAK1 BTRC CASP4 CCR4 CDC7 CHUK CREB1 CXCR1 DICER1 ABCB5 AHNAK2 AQP4 BAMBI BUB1 CASP5 CCR5 CDCA7 CIC CREB3L1 CXCR2 DISP1 ABCB6 AHR AQP7 BAP1 BUB1B CASP6 CCR6 CDH1 CIDEA CREB3L3 CXCR3 DISP2 ABCC1 AHRR AQP9 BATF C17orf53 CASP7 CCR7 CDH13 CIDEB CREB3L4 CXCR4 DKC1 ABCC10 AICDA AR BAX C19orf40 CASP8 CCR8 CDH15 CIRBP CREB5 CXCR5 DKK1 ABCC11 AIFM1 ARAF BBC3 C1orf106 CASP8AP2 CCR9 CDH2 CITED2 CREBBP CXCR6 DKK2 ABCC12 AIMP2 AREG BBS4 C1orf159 CASP9 CCRL2 CDH3 CKB CRK CXXC4 DKK3 ABCC2 AK1 ARHGAP44 BCAR1 C1orf86 CAV1 CCS CDH5 CKLF CRLF2 CXXC5 DKK4 ABCC3 AK2 ARHGEF16 BCAT1 C1QA CAV2 CCT2 CDK1 CKMT1A CRLS1 CYBA DLC1 ABCC4 AK3 ARID1A BCCIP C1S CBL CCT3 CDK16 CKMT2 CRP CYBB DLGAP5 ABCC5 AKAP1 ARID1B BCL10 C3 CBLC CCT4 CDK2 CKS1B CRTAC1 CYCS DLK1 ABCC6 AKR1B1 ARID2 BCL2 C3AR1 CBX3 CCT5 CDK4 CKS2 CRTC2 CYLD DLL1 ABCD1 AKR1C3 ARMC1 BCL2A1 C5 CBX5 CCT6A CDK5 CLCA2 CRY1 CYP19A1 DLL3 ABCD3 AKT1 ARNT BCL2L1 C5AR1 CCBL2 CCT6B CDK5R1 CLCF1 CRYAA CYP1A1 DLL4 -
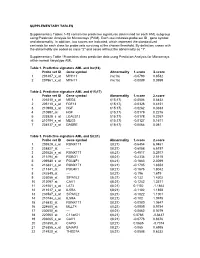
Supplementary Tables 1-18 Contain the Predictive Signatures Determined for Each AML Subgroup Using Prediction Analysis for Microarrays (PAM)
SUPPLEMENTARY TABLES Supplementary Tables 1-18 contain the predictive signatures determined for each AML subgroup using Prediction Analysis for Microarrays (PAM). Each row indicates probe set ID , gene symbol and abnormality. In addition, two scores are indicated, which represent the standardized centroids for each class for probe sets surviving at the chosen threshold. By definition, cases with the abnormality are coded as class "2" and cases without the abnormality as "1". Supplementary Table 19 contains class prediction data using Prediction Analysis for Microarrays within normal karyotype AML. Table 1. Predictive signature AML and inv(16) Probe set ID Gene symbol Abnormality 1.score 2.score 1 201497_x_at MYH11 inv(16) -0.0793 0.8532 2 207961_x_at MYH11 inv(16) -0.0089 0.0959 Table 2. Predictive signature AML and t(15;17) Probe set ID Gene symbol Abnormality 1.score 2.score 1 226210_s_at MEG3 t(15;17) -0.0506 0.6432 2 205110_s_at FGF13 t(15;17) -0.0326 0.4151 3 210998_s_at HGF t(15;17) -0.0262 0.3333 4 210997_at HGF t(15;17) -0.0179 0.2276 5 223828_s_at LGALS12 t(15;17) -0.0178 0.2267 6 210794_s_at MEG3 t(15;17) -0.0127 0.1611 7 204537_s_at GABRE t(15;17) -0.0064 0.081 Table 3. Predictive signature AML and t(8;21) Probe set ID Gene symbol Abnormality 1.score 2.score 1 205529_s_at RUNX1T1 t(8;21) -0.6454 6.9461 2 228827_at --- t(8;21) -0.6058 6.5197 3 205528_s_at RUNX1T1 t(8;21) -0.4917 5.2917 4 213194_at ROBO1 t(8;21) -0.2334 2.5115 5 206940_s_at POU4F1 t(8;21) -0.1883 2.0269 6 216831_s_at RUNX1T1 t(8;21) -0.1705 1.8353 7 211341_at