Download the Full Paper
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A New Hybrid Model Using an Autoregressive Integrated Moving
Am. J. Trop. Med. Hyg., 97(3), 2017, pp. 799–805 doi:10.4269/ajtmh.16-0648 Copyright © 2017 by The American Society of Tropical Medicine and Hygiene A New Hybrid Model Using an Autoregressive Integrated Moving Average and a Generalized Regression Neural Network for the Incidence of Tuberculosis in Heng County, China Wudi Wei,1† Junjun Jiang,1† Lian Gao,2 Bingyu Liang,1 Jiegang Huang,1 Ning Zang,1,3 Chuanyi Ning,1,3 Yanyan Liao,1,3 Jingzhen Lai,1 Jun Yu,1 Fengxiang Qin,1 Hui Chen,4 Jinming Su,1 Li Ye,1* and Hao Liang1,3* 1Guangxi Key Laboratory of AIDS Prevention and Treatment and Guangxi Universities Key Laboratory of Prevention, Guangxi Medical University, 22 Shuangyong Road, Nanning, Guangxi, China; 2Department of Infectious Diseases, Heng County Centers for Disease Control and Prevention, 16 Gongyuan Road, Heng County, China; 3Life Sciences Institute, Guangxi Medical University, 22 Shuangyong Road, Nanning, China; 4Geriatrics Digestion Department of Internal Medicine, The First Affiliated Hospital of Guangxi Medical University, 6 Shuangyong Road, Nanning, China Abstract. It is a daunting task to eradicate tuberculosis completely in Heng County due to a large transient population, human immunodeficiency virus/tuberculosis coinfection, and latent infection. Thus, a high-precision forecasting model can be used for the prevention and control of tuberculosis. In this study, four models including a basic autoregressive integrated moving average (ARIMA) model, a traditional ARIMA–generalized regression neural network (GRNN) model, a basic GRNN model, and a new ARIMA–GRNN hybrid model were used to fit and predict the incidence of tuberculosis. -
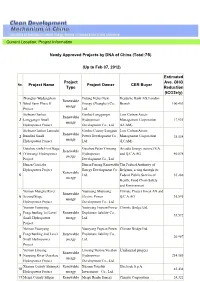
Current Location: Project Information Newly Approved Projects by DNA of China (Total:75) (Up to Feb 07, 2012) Project Name Proje
Current Location: Project Information Newly Approved Projects by DNA of China (Total:75) (Up to Feb 07, 2012) Estimated Project Ave. GHG No. Project Name Project Owner CER Buyer Type Reduction (tCO2e/y) Zhangbei Wudengshan Datang Hebei New Deutsche Bank AG, London Renewable 1 Wind Farm Phase II Energy (Zhangbei) Co., Branch 100,453 energy Project Ltd. Sichuan Ganluo Ganluo Longgangzi Low Carbon Assets Renewable 2 Longgangzi Small Hydropower Management Corporation 17,931 energy Hydropower Project Development Co., Ltd. (LCAM) Sichuan Ganluo Lamodai Ganluo County Longjian Low Carbon Assets Renewable 3 Bundled Small Power Development Co., Management Corporation 25,534 energy Hydropower Project Ltd. (LCAM) Guizhou yinlu First Stage Guizhou Pu'an Yinwang Arcadia Energy (suisse) S.A. Renewable 4 (Yinwang) Hydropower Hydropower and Q.C.A.AG 40,838 energy Project Development Co., Ltd. Hunan Gaojiaba Hunan Furong Renewable The Federal Authority of Hydropower Project Energy Development Co., Belgium, acting through its Renewable 5 Ltd. Federal Public Service of 35,286 energy Health, Food Chain Safety and Environment. Yunnan Mangtie River Yuanyang Shunyang Climate Project Invest AG and Renewable 6 Second Stage Electric Power Q.C.A.AG 35,595 energy Hydropower Project Development Co., Ltd. Yunnan Yuanyang Yuanyang Fuyuan Power Climate Bridge Ltd. Fengchunling 1st Level Renewable Exploiture liability Co., 7 35,592 Small Hydropower energy Ltd. Project Yunnan Yuanyang Yuanyang Fuyuan Power Climate Bridge Ltd. Fengchunling 2nd Level Renewable Exploiture liability Co., 8 26,987 Small Hydropower energy Ltd. Project Yunnan Lincang Lincang Yuntou Yuedian Unilateral project Renewable 9 Nanpeng River Dayakou Hydropower 254,185 energy Hydropower Project Development Co., Ltd. -

Anisotropic Patterns of Liver Cancer Prevalence in Guangxi in Southwest China: Is Local Climate a Contributing Factor?
DOI:http://dx.doi.org/10.7314/APJCP.2015.16.8.3579 Anisotropic Patterns of Liver Cancer Prevalence in Guangxi in Southwest China: Is Local Climate a Contributing Factor? RESEARCH ARTICLE Anisotropic Patterns of Liver Cancer Prevalence in Guangxi in Southwest China: Is Local Climate a Contributing Factor? Wei Deng1&, Long Long2&*, Xian-Yan Tang3, Tian-Ren Huang1, Ji-Lin Li1, Min- Hua Rong1, Ke-Zhi Li1, Hai-Zhou Liu1 Abstract Geographic information system (GIS) technology has useful applications for epidemiology, enabling the detection of spatial patterns of disease dispersion and locating geographic areas at increased risk. In this study, we applied GIS technology to characterize the spatial pattern of mortality due to liver cancer in the autonomous region of Guangxi Zhuang in southwest China. A database with liver cancer mortality data for 1971-1973, 1990-1992, and 2004-2005, including geographic locations and climate conditions, was constructed, and the appropriate associations were investigated. It was found that the regions with the highest mortality rates were central Guangxi with Guigang City at the center, and southwest Guangxi centered in Fusui County. Regions with the lowest mortality rates were eastern Guangxi with Pingnan County at the center, and northern Guangxi centered in Sanjiang and Rongshui counties. Regarding climate conditions, in the 1990s the mortality rate of liver cancer positively correlated with average temperature and average minimum temperature, and negatively correlated with average precipitation. In 2004 through 2005, mortality due to liver cancer positively correlated with the average minimum temperature. Regions of high mortality had lower average humidity and higher average barometric pressure than did regions of low mortality. -

Addition of Clopidogrel to Aspirin in 45 852 Patients with Acute Myocardial Infarction: Randomised Placebo-Controlled Trial
Articles Addition of clopidogrel to aspirin in 45 852 patients with acute myocardial infarction: randomised placebo-controlled trial COMMIT (ClOpidogrel and Metoprolol in Myocardial Infarction Trial) collaborative group* Summary Background Despite improvements in the emergency treatment of myocardial infarction (MI), early mortality and Lancet 2005; 366: 1607–21 morbidity remain high. The antiplatelet agent clopidogrel adds to the benefit of aspirin in acute coronary See Comment page 1587 syndromes without ST-segment elevation, but its effects in patients with ST-elevation MI were unclear. *Collaborators and participating hospitals listed at end of paper Methods 45 852 patients admitted to 1250 hospitals within 24 h of suspected acute MI onset were randomly Correspondence to: allocated clopidogrel 75 mg daily (n=22 961) or matching placebo (n=22 891) in addition to aspirin 162 mg daily. Dr Zhengming Chen, Clinical Trial 93% had ST-segment elevation or bundle branch block, and 7% had ST-segment depression. Treatment was to Service Unit and Epidemiological Studies Unit (CTSU), Richard Doll continue until discharge or up to 4 weeks in hospital (mean 15 days in survivors) and 93% of patients completed Building, Old Road Campus, it. The two prespecified co-primary outcomes were: (1) the composite of death, reinfarction, or stroke; and Oxford OX3 7LF, UK (2) death from any cause during the scheduled treatment period. Comparisons were by intention to treat, and [email protected] used the log-rank method. This trial is registered with ClinicalTrials.gov, number NCT00222573. or Dr Lixin Jiang, Fuwai Hospital, Findings Allocation to clopidogrel produced a highly significant 9% (95% CI 3–14) proportional reduction in death, Beijing 100037, P R China [email protected] reinfarction, or stroke (2121 [9·2%] clopidogrel vs 2310 [10·1%] placebo; p=0·002), corresponding to nine (SE 3) fewer events per 1000 patients treated for about 2 weeks. -

INTEGRATED SAFEGUARDS DATA SHEET CONCEPT STAGE Report No.: AC4020
INTEGRATED SAFEGUARDS DATA SHEET CONCEPT STAGE Report No.: AC4020 Public Disclosure Authorized Date ISDS Prepared/Updated: 12/23/2008 I. BASIC INFORMATION A. Basic Project Data Country: China Project ID: P108627 Project Name: CN - Nanning Urban Environment Task Team Leader: Takuya Kamata Estimated Appraisal Date: March 16, 2009 Estimated Board Date: July 7, 2009 Managing Unit: EASUR Lending Instrument: Specific Investment Loan Public Disclosure Authorized Sector: Sanitation (50%);Solid waste management (25%);Sewerage (25%) Theme: Access to urban services and housing (P) IBRD Amount (US$m.): 150.00 IDA Amount (US$m.): 0.00 GEF Amount (US$m.): 0.00 PCF Amount (US$m.): 0.00 Other financing amounts by source: Borrower 150.00 150.00 B. Project Objectives [from section 2 of PCN] Public Disclosure Authorized The project development objective is to assist the Nanning Municipality in arresting further deterioration of water quality in selected rivers by expanding coverage of wastewater treatment services, carrying out environmental rehabilitation of river courses, and providing technical assistance for development of the integrated mini-river basin management systems. This objective would be achieved by: i) supporting wastewater management through the expansion of treatment capacity for the Jingnan Wastewater Treatment Plant, and the development of new wastewater collection and treatment facilities in five county seat towns; (ii) the rehabilitation and environmental improvement of tributaries of Yongjiang River, including the Fenghuang River, the Liangqing River and Lengtangchong River; and (iii) an innovative technical assistance activity on integrated mini-river basin management to achieve the sustainable social and economic development and improved water ecological environment in the complete Nanning basin area. -

Application of Background Information Database in Drought Monitoring of Guangxi in 2010
Application of Background Information Database in Drought Monitoring of Guangxi in 2010 ∗ Xin Yang1,2, , Weiping Lu1,2, Chaohui Wu1,2, Yuhong Li1,2, and Shiquan Zhong1,2 1 Remote Sensing Application and Test Base of National Satellite Meteorology Centre, Nanning, China, 530022 2 GuangXi Institute of Meteorology, Nanning, China 530022 GuangXi Institute of Meteorology, Nanning, 530022, P.R. China Tel.: +86-771-5875207; Fax: +86-771-5865594 [email protected] Abstract. In this paper, use Nanning city as an example to show application of Background information database in drought monitoring. A near-real time drought monitoring approach is developed using Terra-Moderate Resolution Imaging Spectoradiometer (MODIS) Normalized Difference Vegetation Index (NDVI) and Land Surface Temperature (LST) products. The approach is called Vegetation Temperature Condition Index (VTCI), which integrates land surface reflectance and thermal properties. VTCI is defined as the ratio of LST differ- ences among pixels with a specific NDVI value in a sufficiently large study area; The ground-measured precipitation data from a study area covering Nan- ning in Guangxi , CHINA, are used to validate the drought monitoring ap- proach. Taking the result of drought monitoring in background information of Nanning city ,the area of farmland drought of Mild drought Moderate droughts Severe drought were 223607.2 Ha ,310596.9 Ha and 513.2 Ha. Keywords: Background Information Database, Drought Monitoring, MODIS, Guangxi. 1 Introduction Drought is a normal, recurrent feature of climate. It occurs almost everywhere, although its features vary from region to region. Drought is one of the major envi- ronmental disasters in China, whereas in recent years it have happened in mid and southern of china seriously, so it is very important to detect and monitor drought peri- odically at large scale for decision making. -

EPA-WP-19-F01 Certified Mill List\261\273\310\317\326\244\271
Company Board Type Address TPC Country Province/Region City GUANGXI XIANGSHENG HOUSEHOULD Address:INDUSTRIAL PARK, CHENGZHONG TPC-47 China Guangxi Chongzuo MATERIAL TECHNOLOGY CO., LTD PB TOWN, NINGMING COUNTY, CHOUNGZUO CITY, GUANGXI, P.R. CHINA Guangxi Dongchang Wood Industry Co., Ltd. MDF Address:Industrial Zone, Dasi Town, Qinzhou TPC-47 China Guangxi Qinzhou City, China Zhangzhou Zhongfu New Material Co,Ltd. Thin MDF Address: Shugang Road,Lingang Industrial Park, TPC-47 China Fujian Zhangzhou Yunxiao County,Fujian Province Guigang City Xiangyi Wood Industry Co., Ltd HWPW Address: The southeastern of the intersection of TPC-47 China Guangxi Guigang Chengnan Avenue and West Third Road of Guigang Industrial Park(Jiangnan Park),China FOSHAN NANHAI YINYUAN INSTRUMENTS HWPW Address: Liuchao Shuikou Industrial Zone Lishui TPC-47 China Guangdong Foshan BOARD MATERIAL MANUFACTURE CO., LTD Town Nanhai Dist. Foshan City-528244, Guangdong, China. Fusui Zhongsen Wood Industry Co., Ltd. HWPW Address:No.5 Fenghua Road, Shanxu Town, TPC-47 China Guangxi Chongzuo Fusui County,Chongzuo City, Guangxi Province, China. Guangxi Gaofeng Wuzhou Wood-based Panel Co., Thin MDF&MDF Address:Liutang, Wutang Town, Xingning district, TPC-47 China Guangxi Nanning Ltd. Nanning City, Guangxi, P.R.China Guangxi Fenglin Wooden-Based Panels Co., Ltd. MDF Address:No.26 Mingyang Avenue, Wuxu Town, TPC-47 China Guangxi Nanning Jiangnan District, Nanning City Guangxi Hengxian Liguan Wood-based Panel Co., MDF Adress:Daqiao Development Area, Hengzhou TPC-47 China Guangxi Nanning Ltd. Town, Heng County, Nanning City, Guangxi Guangxi Haolin Wood-Based Panel Co., Ltd. MDF Address: Qinghu Yashan town,Bobai TPC-47 China Guangxi Nanning County,Yulin City, Guangxi Province, China Hubei Province Songzi Hangsen Wood Industry Co., MDF Address: Shugang Road, Lingang Industrial TPC-47 China Hubei Jingzhou Ltd Zone, Songzi City, Hubei Province Guangxi Gaofeng Guishan Wood Based Panel CO., PB Address: Hudie Station, Daguishan Plantation, TPC-47 China Guangxi HeZhou Ltd. -

Copy / Paste the Company's Name of This List Into the Relevant Datafield of Our Webpage by Using the Before Mentioned Link
List of Operators subject to the organic control system according to Commission Regulations (EC) No 1235/2008 Article 11 (3e) and equivalent to (EC) No 834/2007, (EC) No 889/2008. This list has been updated bx Kiwa BCS on 22.04.2021 This list targets at providing information without any legally commitment. Only the Operators' current Certificate is legally binding. For any further questions related to the certification status of any EU-organic Operator certified by Kiwa BCS please contact https://www.kiwa.com/de/de/aktuelle-angelegenheiten/zertifikatssuche/ [email protected] copy / paste the Company's name of this list into the relevant datafield of our webpage by using the before mentioned link. Company Name Location Country Products Status 4 Elementos Industria Barueri BRAZIL Acai, Frozen Foods Certified Alimentos 854 Community Shunli Oil 158403 Hulin City, Heilongjiang CHINA Soybean meal Certified Processing Plant Province Absolute Organix Birnham Park, Gauteng ZA Suedafrika Products as per attachment Certified AÇAÍ AMAZONAS INDUSTRIA OBIDOS, PARA BRAZIL Acai coarse 14% (or special) 84 t; Acai Fine 8% (or Popular) 84 t; Certified E COMERCIO LTDA. Acai powder 1 t; Acai powder 100% pure RWD 1 t; Acerola powder 1 t; acerola powder RWD 1 t; Camu Camu Powder 2 t; Camu Camu Powder RWD 1 t; Camu Camu pulp 0,7 t; Graviola powder 1 t ; Graviola Powdered RWD 1 t; medium acai 11% - 84 t; medium acai 12% - 84 t; Passion fruit powder RWD 1 t; Passion fruit powder 1 t; powder Mango 1 t; powdered cupuaçu 1 t; Powdered cupuaçu RWD 1 t; powdered Mango RWD 1 t; Premix 80/20 Açaí Powder 2 t; Strawberry powder 1 t; Strawberry powder RWD 1 t ADPP Bissorá, Oio GW Guinea-Bissau Cashew nuts, Cashew nuts, raw with shell Certified AGA Armazéns Gerais Araxá Araxá BRAZIL Coffee Beans, Green (3000t) Certified Ltda. -

Smallholder Farming Systems in Southwest China Exploring Key Trends and Innovations for Resilience
Smallholder farming systems in southwest China Exploring key trends and innovations for resilience Yiching Song, Yanyan Zhang, Xin Song and Krystyna Swiderska Country Report Food and agriculture; Climate change Keywords: March 2016 Climate resilience, Smallholder innovation for resilience (SIFOR), small-scale farming, biocultural heritage About the authors Yiching Song; Yanyan Zhang; Xin Song. Centre for Chinese Agricultural Policy (CCAP); Krystyna Swiderska (IIED). [email protected] Produced by IIED’s Natural Resources Group The aim of the Natural Resources Group is to build partnerships, capacity and wise decision-making for fair and sustainable use of natural resources. Our priority in pursuing this purpose is on local control and management of natural resources and other ecosystems. Partner organisation The Centre for Chinese Agricultural Policy (CCAP), Chinese Academy of Science, aims to analyse and help formulate practical and feasible policies related to agricultural R&D, natural resource and environmental issues, rural-urban linkages and sustainable development. It’s Participatory Action Research programme has pioneered China’s first participatory plant breeding programme since 2000. Funding acknowledgement This report and the Smallholder Innovation for Resilience (SIFOR) project have been produced with the financial assistance of the European Union and UKaid from the UK Government. The contents of this document are the sole responsibility of IIED and CCAP and can under no circumstances be regarded as reflecting the position of the European Union or of the UK Government. Published by IIED, March 2016 Song, Y, Zhang, Y, Song, X and Swiderska, K (2016) Smallholder farming systems in southwest China: exploring key trends and innovations for resilience. -

Pharmaceutical Management of Antiretroviral Medicines in Guangxi Province, China: Report of SPS Visits to Treatment and Distribution Sites, December 2008
Pharmaceutical Management of Antiretroviral Medicines in Guangxi Province, China: Report of SPS Visits to Treatment and Distribution Sites, December 2008 Helena Walkowiak Sharri Hollist Connie Osborne Lan Zhang January 2009 Strengthening Pharmaceutical Systems Center for Pharmaceutical Management Management Sciences for Health 4301 N. Fairfax Drive, Suite 400 Arlington, VA 22203 USA Phone: 703.524.6575 Fax: 703.524.7898 E-mail: [email protected] Pharmaceutical Management of Antiretroviral Medicines in Guangxi Province, China: Report of SPS Visits to Treatment and Distribution Sites, December 2008 This report is made possible by the generous support of the American people through the U.S. Agency for International Development (USAID), under the terms of Cooperative Agreement #GHN-A-00-07-00002-00. The contents are the responsibility of Management Sciences for Health and do not necessarily reflect the views of USAID or the United States Government. About SPS The Strengthening Pharmaceutical Systems (SPS) Program strives to build capacity within developing countries to effectively manage all aspects of pharmaceutical systems and services. SPS focuses on improving governance in the pharmaceutical sector, strengthening pharmaceutical management systems and financing mechanisms, containing antimicrobial resistance, and enhancing access to and appropriate use of medicines. Recommended Citation This report may be reproduced if credit is given to SPS. Please use the following citation. Walkowiak, H., S. Hollist, C. Osborne, and L. Zhang. 2009. Pharmaceutical Management of Antiretroviral Medicines in Guangxi Province, China: Report of SPS/WHO Visits to Treatment and Distribution Sites, December 1-10, 2008. Submitted to the U.S. Agency for International Development by the Strengthening Pharmaceutical Systems (SPS) Program. -

Announcement to Australian Securities Exchange Limited (“ASX”) ( ACN 008 624 691)
ViaGOLD Capital Limited (ARBN 070 352 500) Announcement to Australian Securities Exchange Limited (“ASX”) ( ACN 008 624 691) 27 July2015 To Australian Securities Exchange Limited (“ASX”) Company Announcements Office 10th Floor, 20 Bond Street Sydney NSW Re: EXCLUSIVE SALES AND PURCHASE CONTRACT SIGNED - STRENGTHEN THE SUPPORT OF THE VIAGOLD GROUP’S RARE EARTH INDUSTRY CHAIN The Board of Directors is pleased to announce that Zhuhai Hongjie Enterprise Management Consulting Co., Ltd (hereinafter “Hongjie”), an indirect wholly owned subsidiary of ViaGold Capital Limited (ASX Code: VIA) and Guangxi Yuli Minerals Investment Co., Ltd. (hereinafter "Yuli") have entered into an Exclusive Sales and Purchase Contract (hereinafter the “Contract”) on 25 July, 2016. Yuli and its affiliated enterprise Jinshi Mining Development Business in Heng County,Nanning, Guangxi Province are mainly engaging in development, procurement, sales and further processing in refining mineral concentrate. Main product is vanadium titanium iron concentrate. Yuli has discovered abundant overlaid minerals in its tailing ponds. Yuli intends to develop overlaid minerals and is processing to apply for government approval for the rare earth rescue recovery project (hereinafter the "rare earth recovery project matters"). At present, it is forecasted that the reserves of the overlaid rare earth in the tailing ponds in the mining area is about 150,000 tons. The economic value is about 2.3 billion Australian Dollars. It is forecasted that the annual recovery output is expected to be not less than 6,000 tons and is amounted to about 95.6 million Australian Dollars. The principal terms and conditions of the Exclusive Sales and Purchase Contract between Yuli (hereinafter “Party A”) and Hongjie (hereinafter “Party B”) are summarized as follows: ViaGold Capital Limited (Listed in ASX code VIA) Suite 1102, Level 11, 370 Pitt Street Sydney, NSW 2000 +61 411 313 332 +61 2 9283 3933 ARBN 070 352 500 1. -

Application of a Combined Model with Autoregressive Integrated Moving Average (ARIMA) and Generalized Regression Neural Network (GRNN) in Forecasting Hepatitis Incidence in Heng
RESEARCH ARTICLE Application of a Combined Model with Autoregressive Integrated Moving Average (ARIMA) and Generalized Regression Neural Network (GRNN) in Forecasting Hepatitis Incidence in Heng County, China Wudi Wei1☯, Junjun Jiang1☯, Hao Liang1,3, Lian Gao2, Bingyu Liang1, Jiegang Huang1, a11111 Ning Zang1,3, Yanyan Liao1,3, Jun Yu1, Jingzhen Lai1, Fengxiang Qin1, Jinming Su1, Li Ye1,3*, Hui Chen4* 1 Guangxi Key Laboratory of AIDS Prevention and Treatment & Guangxi Universities Key Laboratory of Prevention and Control of Highly Prevalent Disease, School of Public Health, Guangxi Medical University, Nanning 530021, Guangxi, China, 2 Centers for Disease Control and Prevention, Heng County 530300, Guangxi, China, 3 Guangxi Collaborative Innovation Center for Biomedicine, Guangxi Medical Research Center, Guangxi Medical University, Nanning 530021, Guangxi, China, 4 Geriatrics Digestion Department of OPEN ACCESS Internal Medicine, The First Affiliated Hospital of Guangxi Medical University, Nanning 530021, Guangxi, China Citation: Wei W, Jiang J, Liang H, Gao L, Liang B, Huang J, et al. (2016) Application of a Combined ☯ These authors contributed equally to this work. Model with Autoregressive Integrated Moving * [email protected] (HC); [email protected] (LY) Average (ARIMA) and Generalized Regression Neural Network (GRNN) in Forecasting Hepatitis Incidence in Heng County, China. PLoS ONE 11(6): e0156768. doi:10.1371/journal.pone.0156768 Abstract Editor: Sheng-Nan Lu, Kaohsiung Chang Gung Background Memorial Hospital, TAIWAN Hepatitis is a serious public health problem with increasing cases and property damage in Received: January 28, 2016 Heng County. It is necessary to develop a model to predict the hepatitis epidemic that could Accepted: May 19, 2016 be useful for preventing this disease.