The Specificity of Downstream Signaling for A1 and A2AR
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
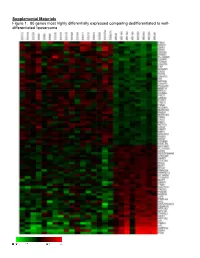
Supplemental Materials Figure 1. 80 Genes Most Highly Differentially Expressed Comparing Dedifferentiated to Well- Differentiated Liposarcoma
Supplemental Materials Figure 1. 80 genes most highly differentially expressed comparing dedifferentiated to well- differentiated liposarcoma Figure 2. Validation of microa CDC2, RACGAP1, FGFR-2, MAD2 and CITED1 200 CDK4 by Liposarcoma Subtype 16 0 12 0 80 Expression40 Level 0 Nl Fat rray data by quantitative RT-P Well Diff 16 0 p16 by Liposarcoma Subtype Dediff 12 0 80 Myxoid Expression40 Level Round Cell 0 12 0 MDM2 by Liposarcoma Subtype 10 0 Pleomorphic Nl Fat 80 60 40 Expression Level 20 RACGAP1 by LiposarcomaWell Diff Subtype 0 70 CR for genes CDK4, MDM2, p16, 60 Dediff 50 Nl Fat 40 30 Myxoid Expression20 Level Well Diff 10 50 0 Round Cell CDC2 by Liposarcoma Subtype Dediff 40 Nl Fat Pleomorphic 30 Myxoid 20 Expression Level Well Diff 10 MAD2L1 by Liposarcoma Subtype Round Cell 60 0 50 Dediff Pleomorphic 40 Nl Fat 30 Myxoid 20 Expression Level Well Diff 10 FGFR-2 by Liposarcoma Subtype 0 Round Cell 200 16 0 Dediff Nl Fat Pleomorphic 12 0 Myxoid 80 Expression Level Well Diff 40 Round Cell 0 Dediff Pleomorphic Nl Fat Myxoid Well Diff CITED1 by Liposarcoma Subtype Round Cell 10 0 80 Dediff Pleomorphic 60 40 Myxoid Expression Level 20 0 Round Cell Nl Fat Pleomorphic Well Diff Dediff Myxoid Round Cell Pleomorphic Table 1. 142 Gene classifier for liposarcoma subtype The genes used for each pair wise subtype comparison are grouped together. The flag column indicates which genes are unique to each subtype comparison. The values show the mean expression levels (actually the mean of the log expression levels was computed and than transformed back to absolute expression level). -

Supplemental Material
Supplemental Table B ARGs in alphabetical order Symbol Title 3 months 6 months 9 months 12 months 23 months ANOVA Direction Category 38597 septin 2 1557 ± 44 1555 ± 44 1579 ± 56 1655 ± 26 1691 ± 31 0.05219 up Intermediate 0610031j06rik kidney predominant protein NCU-G1 491 ± 6 504 ± 14 503 ± 11 527 ± 13 534 ± 12 0.04747 up Early Adult 1G5 vesicle-associated calmodulin-binding protein 662 ± 23 675 ± 17 629 ± 16 617 ± 20 583 ± 26 0.03129 down Intermediate A2m alpha-2-macroglobulin 262 ± 7 272 ± 8 244 ± 6 290 ± 7 353 ± 16 0.00000 up Midlife Aadat aminoadipate aminotransferase (synonym Kat2) 180 ± 5 201 ± 12 223 ± 7 244 ± 14 275 ± 7 0.00000 up Early Adult Abca2 ATP-binding cassette, sub-family A (ABC1), member 2 958 ± 28 1052 ± 58 1086 ± 36 1071 ± 44 1141 ± 41 0.05371 up Early Adult Abcb1a ATP-binding cassette, sub-family B (MDR/TAP), member 1A 136 ± 8 147 ± 6 147 ± 13 155 ± 9 185 ± 13 0.01272 up Midlife Acadl acetyl-Coenzyme A dehydrogenase, long-chain 423 ± 7 456 ± 11 478 ± 14 486 ± 13 512 ± 11 0.00003 up Early Adult Acadvl acyl-Coenzyme A dehydrogenase, very long chain 426 ± 14 414 ± 10 404 ± 13 411 ± 15 461 ± 10 0.01017 up Late Accn1 amiloride-sensitive cation channel 1, neuronal (degenerin) 242 ± 10 250 ± 9 237 ± 11 247 ± 14 212 ± 8 0.04972 down Late Actb actin, beta 12965 ± 310 13382 ± 170 13145 ± 273 13739 ± 303 14187 ± 269 0.01195 up Midlife Acvrinp1 activin receptor interacting protein 1 304 ± 18 285 ± 21 274 ± 13 297 ± 21 341 ± 14 0.03610 up Late Adk adenosine kinase 1828 ± 43 1920 ± 38 1922 ± 22 2048 ± 30 1949 ± 44 0.00797 up Early -

Molecular Dissection of G-Protein Coupled Receptor Signaling and Oligomerization
MOLECULAR DISSECTION OF G-PROTEIN COUPLED RECEPTOR SIGNALING AND OLIGOMERIZATION BY MICHAEL RIZZO A Dissertation Submitted to the Graduate Faculty of WAKE FOREST UNIVERSITY GRADUATE SCHOOL OF ARTS AND SCIENCES in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY Biology December, 2019 Winston-Salem, North Carolina Approved By: Erik C. Johnson, Ph.D. Advisor Wayne E. Pratt, Ph.D. Chair Pat C. Lord, Ph.D. Gloria K. Muday, Ph.D. Ke Zhang, Ph.D. ACKNOWLEDGEMENTS I would first like to thank my advisor, Dr. Erik Johnson, for his support, expertise, and leadership during my time in his lab. Without him, the work herein would not be possible. I would also like to thank the members of my committee, Dr. Gloria Muday, Dr. Ke Zhang, Dr. Wayne Pratt, and Dr. Pat Lord, for their guidance and advice that helped improve the quality of the research presented here. I would also like to thank members of the Johnson lab, both past and present, for being valuable colleagues and friends. I would especially like to thank Dr. Jason Braco, Dr. Jon Fisher, Dr. Jake Saunders, and Becky Perry, all of whom spent a great deal of time offering me advice, proofreading grants and manuscripts, and overall supporting me through the ups and downs of the research process. Finally, I would like to thank my family, both for instilling in me a passion for knowledge and education, and for their continued support. In particular, I would like to thank my wife Emerald – I am forever indebted to you for your support throughout this process, and I will never forget the sacrifices you made to help me get to where I am today. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid
The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome Annette K. Brenner, Elise Aasebø, Maria Hernandez-Valladares, Frode Selheim, Frode Berven, Ida-Sofie Grønningsæter, Sushma Bartaula-Brevik and Øystein Bruserud Supplementary Material S2 of S31 Table S1. Detailed information about the 68 AML patients included in the study. # of blasts Viability Proliferation Cytokine Viable cells Change in ID Gender Age Etiology FAB Cytogenetics Mutations CD34 Colonies (109/L) (%) 48 h (cpm) secretion (106) 5 weeks phenotype 1 M 42 de novo 241 M2 normal Flt3 pos 31.0 3848 low 0.24 7 yes 2 M 82 MF 12.4 M2 t(9;22) wt pos 81.6 74,686 low 1.43 969 yes 3 F 49 CML/relapse 149 M2 complex n.d. pos 26.2 3472 low 0.08 n.d. no 4 M 33 de novo 62.0 M2 normal wt pos 67.5 6206 low 0.08 6.5 no 5 M 71 relapse 91.0 M4 normal NPM1 pos 63.5 21,331 low 0.17 n.d. yes 6 M 83 de novo 109 M1 n.d. wt pos 19.1 8764 low 1.65 693 no 7 F 77 MDS 26.4 M1 normal wt pos 89.4 53,799 high 3.43 2746 no 8 M 46 de novo 26.9 M1 normal NPM1 n.d. n.d. 3472 low 1.56 n.d. no 9 M 68 MF 50.8 M4 normal D835 pos 69.4 1640 low 0.08 n.d. -

Predicting Coupling Probabilities of G-Protein Coupled Receptors Gurdeep Singh1,2,†, Asuka Inoue3,*,†, J
Published online 30 May 2019 Nucleic Acids Research, 2019, Vol. 47, Web Server issue W395–W401 doi: 10.1093/nar/gkz392 PRECOG: PREdicting COupling probabilities of G-protein coupled receptors Gurdeep Singh1,2,†, Asuka Inoue3,*,†, J. Silvio Gutkind4, Robert B. Russell1,2,* and Francesco Raimondi1,2,* 1CellNetworks, Bioquant, Heidelberg University, Im Neuenheimer Feld 267, 69120 Heidelberg, Germany, 2Biochemie Zentrum Heidelberg (BZH), Heidelberg University, Im Neuenheimer Feld 328, 69120 Heidelberg, Germany, 3Graduate School of Pharmaceutical Sciences, Tohoku University, Sendai, Miyagi 980-8578, Japan and 4Department of Pharmacology and Moores Cancer Center, University of California, San Diego, La Jolla, CA 92093, USA Received February 10, 2019; Revised April 13, 2019; Editorial Decision April 24, 2019; Accepted May 01, 2019 ABSTRACT great use in tinkering with signalling pathways in living sys- tems (5). G-protein coupled receptors (GPCRs) control multi- Ligand binding to GPCRs induces conformational ple physiological states by transducing a multitude changes that lead to binding and activation of G-proteins of extracellular stimuli into the cell via coupling to situated on the inner cell membrane. Most of mammalian intra-cellular heterotrimeric G-proteins. Deciphering GPCRs couple with more than one G-protein giving each which G-proteins couple to each of the hundreds receptor a distinct coupling profile (6) and thus specific of GPCRs present in a typical eukaryotic organism downstream cellular responses. Determining these coupling is therefore critical to understand signalling. Here, profiles is critical to understand GPCR biology and phar- we present PRECOG (precog.russelllab.org): a web- macology. Despite decades of research and hundreds of ob- server for predicting GPCR coupling, which allows served interactions, coupling information is still missing for users to: (i) predict coupling probabilities for GPCRs many receptors and sequence determinants of coupling- specificity are still largely unknown. -

Quantification of Cardiovascular Disease Biomarkers in Human
proteomes Article Quantification of Cardiovascular Disease Biomarkers in Human Platelets by Targeted Mass Spectrometry Sebastian Malchow ID , Christina Loosse, Albert Sickmann and Christin Lorenz * Leibniz-Institut für Analytische Wissenschaften-ISAS-e.V., 44139 Dortmund, Germany; [email protected] (S.M.); [email protected] (C.L.); [email protected] (A.S.) * Correspondence: [email protected]; Tel.: +49-231-1392-289 Received: 3 October 2017; Accepted: 13 November 2017; Published: 15 November 2017 Abstract: Platelets are known to be key players in thrombosis and hemostasis, contributing to the genesis and progression of cardiovascular diseases. Due to their pivotal role in human physiology and pathology, platelet function is regulated tightly by numerous factors which have either stimulatory or inhibitory effects. A variety of factors, e.g., collagen, fibrinogen, ADP, vWF, thrombin, and thromboxane promote platelet adhesion and aggregation by utilizing multiple intracellular signal cascades. To quantify platelet proteins for this work, a targeted proteomics workflow was applied. In detail, platelets are isolated and lyzed, followed by a tryptic protein digest. Subsequently, a mix of stable isotope-labeled peptides of interesting biomarker proteins in concentrations ranging from 0.1 to 100 fmol is added to 3 µg digest. These peptides are used as an internal calibration curve to accurately quantify endogenous peptides and corresponding proteins in a pooled platelet reference sample by nanoLC-MS/MS with parallel reaction monitoring. In order to assure a valid quantification, limit of detection (LOD) and limit of quantification (LOQ), as well as linear range, were determined. This quantification of platelet activation and proteins by targeted mass spectrometry may enable novel diagnostic strategies in the detection and prevention of cardiovascular diseases. -

Genomic Instability and DNA Ploidy Are Linked to DNA Copy Number Aberrations of 8P23 and 22Q11.23 in Gastric Cancers
333-339.qxd 15/7/2010 11:08 Ì ™ÂÏ›‰·333 INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 26: 333-339, 2010 333 Genomic instability and DNA ploidy are linked to DNA copy number aberrations of 8p23 and 22q11.23 in gastric cancers SHIGETO KAWAUCHI1, TOMOKO FURUAY1, TETSUJI UCHIYAMA2, ATSUSHI ADACHI2, TAKAE OKADA1 MOTONAO NAKAO1, ATSUNORI OGA1, KENICHIRO UCHIDA1 and KOHSUKE SASAKI1 1Department of Pathology, Yamaguchi University Graduate School of Medicine, Ube 755-8505; 2Department of Surgery, Iwakuni Medical Center, Iwakuni 740-0021, Japan Received April 8, 2010; Accepted June 2, 2010 DOI: 10.3892/ijmm_00000470 Abstract. The close relationship between chromosomal Introduction instability (CIN) and aneuploidy has been reported. The purpose of this study was to identify genomic aberrations Cancer progression is accompanied by the accumulation of present with CIN and aneuploidy in gastric cancers. FISH genetic alterations in the genes controlling cell proliferation and image cytometry were applied to 27 sporadic gastric and death. Indeed, some genomic abnormalities, ranging adenocarcinomas to identify CIN-positive tumors and to from point mutations to chromosomal aberrations, are determine DNA ploidy, respectively. In addition, array-based detected in virtually all cancers, including gastric cancers. comparative genomic hybridization was used to identify Although the biological characteristics of cancers greatly bacterial artificial chromosome clones that displayed vary from case to case, they are primarily affected by genomic differences in the frequency of copy number aberrations alterations. In general, cancer cells inherently take on genomic between CIN-positive and CIN-negative tumors, and instability that is conceptually divided into microsatellite between aneuploid and diploid tumors. There were many instability (MIN) and chromosomal instability (CIN) (1,2). -

G-Protein ␥-Complex Is Crucial for Efficient Signal Amplification in Vision
The Journal of Neuroscience, June 1, 2011 • 31(22):8067–8077 • 8067 Cellular/Molecular G-Protein ␥-Complex Is Crucial for Efficient Signal Amplification in Vision Alexander V. Kolesnikov,1 Loryn Rikimaru,2 Anne K. Hennig,1 Peter D. Lukasiewicz,1 Steven J. Fliesler,4,5,6,7 Victor I. Govardovskii,8 Vladimir J. Kefalov,1 and Oleg G. Kisselev2,3 1Department of Ophthalmology and Visual Sciences, Washington University School of Medicine, St. Louis, Missouri 63110, Departments of 2Ophthalmology and 3Biochemistry and Molecular Biology, Saint Louis University School of Medicine, Saint Louis, Missouri 63104, 4Research Service, Veterans Administration Western New York Healthcare System, and Departments of 5Ophthalmology (Ross Eye Institute) and 6Biochemistry, University at Buffalo/The State University of New York (SUNY), and 7SUNY Eye Institute, Buffalo, New York 14215, and 8Sechenov Institute for Evolutionary Physiology and Biochemistry, Russian Academy of Sciences, Saint Petersburg 194223, Russia A fundamental question of cell signaling biology is how faint external signals produce robust physiological responses. One universal mechanism relies on signal amplification via intracellular cascades mediated by heterotrimeric G-proteins. This high amplification system allows retinal rod photoreceptors to detect single photons of light. Although much is now known about the role of the ␣-subunit of the rod-specific G-protein transducin in phototransduction, the physiological function of the auxiliary ␥-complex in this process remains a mystery. Here, we show that elimination of the transducin ␥-subunit drastically reduces signal amplification in intact mouse rods. The consequence is a striking decline in rod visual sensitivity and severe impairment of nocturnal vision. Our findings demonstrate that transducin ␥-complex controls signal amplification of the rod phototransduction cascade and is critical for the ability of rod photoreceptors to function in low light conditions. -

G Protein Alpha Inhibitor 1 (GNAI1) Rabbit Polyclonal Antibody Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA335036 G protein alpha inhibitor 1 (GNAI1) Rabbit Polyclonal Antibody Product data: Product Type: Primary Antibodies Applications: WB Recommended Dilution: WB Reactivity: Human, Mouse, Rat Host: Rabbit Isotype: IgG Clonality: Polyclonal Immunogen: The immunogen for anti-GNAI1 antibody: synthetic peptide directed towards the middle region of human GNAI1. Synthetic peptide located within the following region: YQLNDSAAYYLNDLDRIAQPNYIPTQQDVLRTRVKTTGIVETHFTFKDLH Formulation: Liquid. Purified antibody supplied in 1x PBS buffer with 0.09% (w/v) sodium azide and 2% sucrose. Note that this product is shipped as lyophilized powder to China customers. Purification: Affinity Purified Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 40 kDa Gene Name: G protein subunit alpha i1 Database Link: NP_002060 Entrez Gene 14677 MouseEntrez Gene 25686 RatEntrez Gene 2770 Human P63096 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 4 G protein alpha inhibitor 1 (GNAI1) Rabbit Polyclonal Antibody – TA335036 Background: Guanine nucleotide-binding proteins (G proteins) form a large family of signal-transducing molecules. They are found as heterotrimers made up of alpha, beta, and gamma subunits. Members of the G protein family have been characterized most extensively on the basis of the alpha subunit, which binds guanine nucleotide, is capable of hydrolyzing GTP, and interacts with specific receptor and effector molecules. -

GNAT1 (NM 144499) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC221246 GNAT1 (NM_144499) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: GNAT1 (NM_144499) Human Tagged ORF Clone Tag: Myc-DDK Symbol: GNAT1 Synonyms: CSNB1G; CSNBAD3; GBT1; GNATR Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RC221246 representing NM_144499 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGGGGGCTGGGGCCAGTGCTGAGGAGAAGCACTCCAGGGAGCTGGAAAAGAAGCTGAAAGAGGACGCTG AGAAGGATGCTCGAACCGTGAAGCTGCTGCTTCTGGGTGCCGGTGAGTCCGGGAAGAGCACCATCGTCAA GCAGATGAAGATTATCCACCAGGACGGGTACTCGCTGGAAGAGTGCCTCGAGTTTATCGCCATCATCTAC GGCAACACGTTGCAGTCCATCCTGGCCATCGTACGCGCCATGACCACACTCAACATCCAGTACGGAGACT CTGCACGCCAGGACGACGCCCGGAAGCTGATGCACATGGCAGACACTATCGAGGAGGGCACGATGCCCAA GGAGATGTCGGACATCATCCAGCGGCTGTGGAAGGACTCCGGTATCCAGGCCTGTTTTGAGCGCGCCTCG GAGTACCAGCTCAACGACTCGGCGGGCTACTACCTCTCCGACCTGGAGCGCCTGGTAACCCCGGGCTACG TGCCCACCGAGCAGGACGTGCTGCGCTCGCGAGTCAAGACCACTGGCATCATCGAGACGCAGTTCTCCTT CAAGGATCTCAACTTCCGGATGTTCGATGTGGGCGGGCAGCGCTCGGAGCGCAAGAAGTGGATCCACTGC TTCGAGGGCGTGACCTGCATCATCTTCATCGCGGCGCTGAGCGCCTACGACATGGTGCTAGTGGAGGACG ACGAAGTGAACCGCATGCACGAGAGCCTGCACCTGTTCAACAGCATCTGCAACCACCGCTACTTCGCCAC GACGTCCATCGTGCTCTTCCTTAACAAGAAGGACGTCTTCTTCGAGAAGATCAAGAAGGCGCACCTCAGC ATCTGTTTCCCGGACTACGATGGACCCAACACCTACGAGGACGCCGGCAACTACATCAAGGTGCAGTTCC -

G Protein-Coupled Receptors
G PROTEIN-COUPLED RECEPTORS Overview:- The completion of the Human Genome Project allowed the identification of a large family of proteins with a common motif of seven groups of 20-24 hydrophobic amino acids arranged as α-helices. Approximately 800 of these seven transmembrane (7TM) receptors have been identified of which over 300 are non-olfactory receptors (see Frederikson et al., 2003; Lagerstrom and Schioth, 2008). Subdivision on the basis of sequence homology allows the definition of rhodopsin, secretin, adhesion, glutamate and Frizzled receptor families. NC-IUPHAR recognizes Classes A, B, and C, which equate to the rhodopsin, secretin, and glutamate receptor families. The nomenclature of 7TM receptors is commonly used interchangeably with G protein-coupled receptors (GPCR), although the former nomenclature recognises signalling of 7TM receptors through pathways not involving G proteins. For example, adiponectin and membrane progestin receptors have some sequence homology to 7TM receptors but signal independently of G-proteins and appear to reside in membranes in an inverted fashion compared to conventional GPCR. Additionally, the NPR-C natriuretic peptide receptor has a single transmembrane domain structure, but appears to couple to G proteins to generate cellular responses. The 300+ non-olfactory GPCR are the targets for the majority of drugs in clinical usage (Overington et al., 2006), although only a minority of these receptors are exploited therapeutically. Signalling through GPCR is enacted by the activation of heterotrimeric GTP-binding proteins (G proteins), made up of α, β and γ subunits, where the α and βγ subunits are responsible for signalling. The α subunit (tabulated below) allows definition of one series of signalling cascades and allows grouping of GPCRs to suggest common cellular, tissue and behavioural responses.