Methods for Detection of Phytophthora Fragariae Var. Rubi on Raspberry
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phytopythium: Molecular Phylogeny and Systematics
Persoonia 34, 2015: 25–39 www.ingentaconnect.com/content/nhn/pimj RESEARCH ARTICLE http://dx.doi.org/10.3767/003158515X685382 Phytopythium: molecular phylogeny and systematics A.W.A.M. de Cock1, A.M. Lodhi2, T.L. Rintoul 3, K. Bala 3, G.P. Robideau3, Z. Gloria Abad4, M.D. Coffey 5, S. Shahzad 6, C.A. Lévesque 3 Key words Abstract The genus Phytopythium (Peronosporales) has been described, but a complete circumscription has not yet been presented. In the present paper we provide molecular-based evidence that members of Pythium COI clade K as described by Lévesque & de Cock (2004) belong to Phytopythium. Maximum likelihood and Bayesian LSU phylogenetic analysis of the nuclear ribosomal DNA (LSU and SSU) and mitochondrial DNA cytochrome oxidase Oomycetes subunit 1 (COI) as well as statistical analyses of pairwise distances strongly support the status of Phytopythium as Oomycota a separate phylogenetic entity. Phytopythium is morphologically intermediate between the genera Phytophthora Peronosporales and Pythium. It is unique in having papillate, internally proliferating sporangia and cylindrical or lobate antheridia. Phytopythium The formal transfer of clade K species to Phytopythium and a comparison with morphologically similar species of Pythiales the genera Pythium and Phytophthora is presented. A new species is described, Phytopythium mirpurense. SSU Article info Received: 28 January 2014; Accepted: 27 September 2014; Published: 30 October 2014. INTRODUCTION establish which species belong to clade K and to make new taxonomic combinations for these species. To achieve this The genus Pythium as defined by Pringsheim in 1858 was goal, phylogenies based on nuclear LSU rRNA (28S), SSU divided by Lévesque & de Cock (2004) into 11 clades based rRNA (18S) and mitochondrial DNA cytochrome oxidase1 (COI) on molecular systematic analyses. -

Molecular Identification and Detection of Phytophthora Species on Some Important Mediterranean Plants Including Sweet Chestnut
For. Snow Landsc. Res. 76, 3: 351–356 (2001) 351 Molecular identification and detection of Phytophthora species on some important Mediterranean plants including sweet chestnut Santa Olga Cacciola1, Naomi A. Williams2, David E. L. Cooke2 and James M. Duncan2 1 Dipartimento di Scienze Entomologiche Fitopatologiche, Microbiologiche Agrarie e Zootecniche – Sezione di Patologia Vegetale e Microbiologia Agraria 90128 Palermo, University of Palermo, Italy [email protected] 2 Scottish Crop Research Institute, Invergowrie, Dundee, DD2 5DA, Scotland, U.K. [email protected] Abstract PCR amplification of the internal transcribed spacer regions and 5.8S gene of rDNA and sequencing or restriction digestion of the resultant product were used to confirm the identities of a collection of Phytophthora isolates from a variety of hosts in Italy. All isolates were of previously described species, but a number of new host-pathogen combinations of potential economic import ance were found, including P. palmivora on olive (Olea europaea) trees. Three Phyto ph - thora isolates from chestnut (Castanea sativa) trees were identified as P. cambivora, a known pathogen of chestnut. Existing, but previously unpublished, rapid, sensitive, DNA-based assays for P. cambivora and P. cinnamomi are described. In combination with similar diagnostics for other species such as P. cactorum, these assays could be used to detect nearly all important Phytophthora species known to occur in chestnut. Keywords: Phytophthora, ITS-RFLP, specific primers, diagnosis, Castanea sativa, Italy 1 Introduction The genus Phytophthora comprises approximately 60 species, nearly all of which are pathogens that damage plants and some of which are among the most important pathogens worldwide (ERWIN and RIBEIRO 1996). -

Economic Cost of Invasive Non-Native Species on Great Britain F
The Economic Cost of Invasive Non-Native Species on Great Britain F. Williams, R. Eschen, A. Harris, D. Djeddour, C. Pratt, R.S. Shaw, S. Varia, J. Lamontagne-Godwin, S.E. Thomas, S.T. Murphy CAB/001/09 November 2010 www.cabi.org 1 KNOWLEDGE FOR LIFE The Economic Cost of Invasive Non-Native Species on Great Britain Acknowledgements This report would not have been possible without the input of many people from Great Britain and abroad. We thank all the people who have taken the time to respond to the questionnaire or to provide information over the phone or otherwise. Front Cover Photo – Courtesy of T. Renals Sponsors The Scottish Government Department of Environment, Food and Rural Affairs, UK Government Department for the Economy and Transport, Welsh Assembly Government FE Williams, R Eschen, A Harris, DH Djeddour, CF Pratt, RS Shaw, S Varia, JD Lamontagne-Godwin, SE Thomas, ST Murphy CABI Head Office Nosworthy Way Wallingford OX10 8DE UK and CABI Europe - UK Bakeham Lane Egham Surrey TW20 9TY UK CABI Project No. VM10066 2 The Economic Cost of Invasive Non-Native Species on Great Britain Executive Summary The impact of Invasive Non-Native Species (INNS) can be manifold, ranging from loss of crops, damaged buildings, and additional production costs to the loss of livelihoods and ecosystem services. INNS are increasingly abundant in Great Britain and in Europe generally and their impact is rising. Hence, INNS are the subject of considerable concern in Great Britain, prompting the development of a Non-Native Species Strategy and the formation of the GB Non-Native Species Programme Board and Secretariat. -

Raspberry Breeding
RASPBERRY BREEDING JULIE GRAHAM and NIKKI JENNINGS WORLD PRODUCTION Raspberries are grown in many parts of the world with production estimated at 482, 763 MT (2005) (http:/ FAOSTAT.FAO.ORG). Europe is estimated to produce around half of all production (Rubus idaeus L.). This is an important high-value horticultural industry in many European countries, providing employment directly in agriculture, and indirectly in food processing and confectionary. Most raspberry production is concentrated in the northern and central European countries, although there is an increasing interest in growing cane fruits in southern Europe e.g. in Greece, Italy, Portugal and Spain. In many production areas, the fruit is grown for the fresh market, but in central Europe e.g. Poland, Hungary and Serbia, a high proportion of the crop is destined for processing. Major regions of production in North America include the Pacific North-West, California, Texas and Arkansas, as well as regions in New York, Michigan, Pennsylvania and Ohio. Chile, Argentina and Guatemala also have extensive production. In Europe in particular, there has been increased interest in sale of raspberry fruits harvested from 'organic production' - farming based on methods relying entirely on crop rotation and avoidance of pesticide application (except certain substances currently permitted by the national regulatory authority for organic farming). However, with woody perennial crops the difficulties of maintaining healthy productive plantations over many years are profound and it is too early to judge the overall success of these ventures in Rubus cane fruits. Increasing popularity of autumn-fruiting raspberries, in which Raspberry Breeding late season fruit is harvested from berries forming on the upper nodes of primocanes (Jennings and Brennan 2002), has extended the production season and the period of attack of some foliar and cane pests. -

The Phytophthora Cactorum Genome Provides Insights Into The
www.nature.com/scientificreports Corrected: Author Correction OPEN The Phytophthora cactorum genome provides insights into the adaptation to host defense Received: 30 October 2017 Accepted: 12 April 2018 compounds and fungicides Published online: 25 April 2018 Min Yang1,2, Shengchang Duan1,3, Xinyue Mei1,2, Huichuan Huang 1,2, Wei Chen1,4, Yixiang Liu1,2, Cunwu Guo1,2, Ting Yang1,2, Wei Wei1,2, Xili Liu5, Xiahong He1,2, Yang Dong1,4 & Shusheng Zhu1,2 Phytophthora cactorum is a homothallic oomycete pathogen, which has a wide host range and high capability to adapt to host defense compounds and fungicides. Here we report the 121.5 Mb genome assembly of the P. cactorum using the third-generation single-molecule real-time (SMRT) sequencing technology. It is the second largest genome sequenced so far in the Phytophthora genera, which contains 27,981 protein-coding genes. Comparison with other Phytophthora genomes showed that P. cactorum had a closer relationship with P. parasitica, P. infestans and P. capsici. P. cactorum has similar gene families in the secondary metabolism and pathogenicity-related efector proteins compared with other oomycete species, but specifc gene families associated with detoxifcation enzymes and carbohydrate-active enzymes (CAZymes) underwent expansion in P. cactorum. P. cactorum had a higher utilization and detoxifcation ability against ginsenosides–a group of defense compounds from Panax notoginseng–compared with the narrow host pathogen P. sojae. The elevated expression levels of detoxifcation enzymes and hydrolase activity-associated genes after exposure to ginsenosides further supported that the high detoxifcation and utilization ability of P. cactorum play a crucial role in the rapid adaptability of the pathogen to host plant defense compounds and fungicides. -

Phytophthora Sojae Infecting Soybean: Pathotype Diversity, New Sources
South Dakota State University Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange Theses and Dissertations 2017 Phytophthora Sojae Infecting Soybean: Pathotype Diversity, New Sources of Resistance and Interaction with the Soybean Cyst Nematode Rawnaq Nazneen Chowdhury South Dakota State University Follow this and additional works at: http://openprairie.sdstate.edu/etd Part of the Agricultural Science Commons, and the Agronomy and Crop Sciences Commons Recommended Citation Chowdhury, Rawnaq Nazneen, "Phytophthora Sojae Infecting Soybean: Pathotype Diversity, New Sources of Resistance and Interaction with the Soybean Cyst Nematode" (2017). Theses and Dissertations. 1186. http://openprairie.sdstate.edu/etd/1186 This Dissertation - Open Access is brought to you for free and open access by Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange. For more information, please contact [email protected]. PHYTOPHTHORA SOJAE INFECTING SOYBEAN: PATHOTYPE DIVERSITY, NEW SOURCES OF RESISTANCE AND INTERACTION WITH THE SOYBEAN CYST NEMATODE BY RAWNAQ NAZNEEN CHOWDHURY A dissertation submitted in partial fulfillment of the requirements for the Doctor of Philosophy Major in Plant Science South Dakota State University 2017 iii I would like to dedicate this thesis to my family; my mother Bilquis Alam Chowdhury, my father Raisul Alam Chowdhury, my sister Reema Najma Chowdhury and my brother Late Arif Alam Chowdhury. They have always encouraged me to pursue my passions. I am forever grateful for their never-ending love and support. iv ACKNOWLEDGMENTS With faith and gratitude to the almighty, I would like to express my earnest thanks to give me an opportunity to make my life meaningful in the world. -

HDC Factsheet 02/07 F.Indd
Factsheet 02/07 Horticultural Bradbourne House Development East Malling Cane fruit Council Kent ME19 6DZ T: 01732 848383 F: 01732 848498 E: [email protected] Phytophthora root rot of raspberry and other cane fruits by Scott Raffle and Janet Allen, ADAS This factsheet provides information on the biology of Phytophthora fragariae var. rubi and the effect it has on raspberry and other cane fruit crops. It summarises techniques that should be employed both to avoid and contain infection and offers the currently approved control measures. Background being Phytophthora fragariae var. rubi uncommon for plantations to die out (P. fragariae var. rubi). The disease was completely within three years of initial Phytophthora root rot of raspberry and first reported in raspberry plantations in infection occurring. The disease led other cane fruit crops brings about Scotland in 1937. Further reports were to significant reductions in total yields root death, leading to die back of both made in North America in the 1950’s being produced by raspberry planta- the floricanes (fruiting canes) and pri- and 1960’s, but it wasn’t until the mid tions in Scotland in the 1980’s and mocanes. The root rot can be caused 1970’s that serious outbreaks began to 1990’s, and made a significant con- by a number of species of the soil- occur in the United Kingdom, leading tribution to the rapid decline of rasp- borne fungi Phytophthora. The most to significant loss of plantations in berry production in Scotland during important is now recognised as both Scotland and England. It is not that period (Figure 1). -

Gene-For-Gene Relationships Between Strawberry and the Causal Agent of Red Stele Root Rot, Phytophthorafragariae Var
Gene-for-gene relationships between strawberry and the causal agent of red stele root rot, Phytophthorafragariae var . fragariae. W.E. van de Weg 1360 ^fâTn) , Z2?3 Gene-for-gene relationships between strawberry and the causal agent of red stele root rot, Phytophthorafragariae var. fragariae. Gen-om-gen relaties tussen aardbei en de veroorzaker van roodwortelrot, Phytophthorafragariae var. fragariae. promotor:prof. dr. ir. J.E. Parlevliet hoogleraar in de plantenveredeling co-promotor:dr. ir. A.P.M, den Nijs Hoofd afdeling Groenten en Fruitgewassen CPRO-DLO 3 /OMOÔZO», 'L^ -. W.E. van de Weg Gene-for-gene relationships between strawberry and the causal agent of red stele root rot, Phytophthorafragariae var; fragariae Proefschrift ter verkrijging van de graad van doctor op gezag van de rector magnificus van de Landbouwuniversiteit Wageningen prof. dr. ir. C.M. Karssen, in het openbaar te verdedigen op maandag 23jun i 1997 des namiddags te half twee in de Aula van de Lanbouwuniversiteit te Wageningen j ,4 -' ' '• <K CIP-DATA KONINKLIJKE BIBLIOTHEEK, DEN HAAG Van de Weg, W.E. Gene-for-gene relationships between strawberry and the causal agent of red stele root rot, Phytophthorafragariae var.fragariae I W.E. van de Weg. - [S.I.: s.n] Thesis Wageningen - With ref. - With summary in Dutch ISBN 90-5485-707-2 Subject headings: Fragariaspp., Phytophthorafragariae, strawberry, genetics, disease resistance ®199 7 W.E. van de Weg. No part of this publication may be reproduced, stored in a retrieval system or transmitted in any form by any means, electronic, mechanical, photocopying, recording or otherwise, without the permission of the author, or, where appropriate, of the copyright holder. -
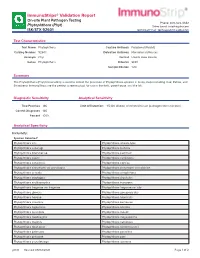
Immunostrips® Validation Report
ImmunoStrips® Validation Report On-site Plant Pathogen Testing Phone: 800-622-4342 Phytophthora (Phyt) Sales Email: [email protected] ISK/STX 92601 Technical Email: [email protected] Test Characteristics Test Name Phytophthora Capture Antibody Polyclonal (Rabbit) Catalog Number 92601 Detection Antibody Monoclonal (Mouse) Acronym Phyt Format Lateral Flow Device Genus Phytophthora Diluents SEB1 Sample Dilution 1:20 Summary The Phytophthora (Phyt) ImmunoStrip is used to detect the presence of Phytophthora species in many crops including Oak, Potato, and Strawberry. ImmunoStrips are the perfect screening tool for use in the field, greenhouse, and the lab. Diagnostic Sensitivity Analytical Sensitivity True Positives 146 Limit of Detection: 1:5,120 dilution of infected tissue (pathogen titer unknown) Correct Diagnoses 146 Percent 100% Analytical Specificity Inclusivity: Species Detected1: Phytophthora alni Phytophthora alticola-type Phytophthora asparagi Phytophthora bisheria Phytophthora boehmeriae Phytophthora cactorum Phytophthora cajani Phytophthora cambivora Phytophthora canalensis Phytophthora capsica Phytophthora cinnamomi var parvispora Phytophthora cinnamomi var robiniae Phytophthora citricola Phytophthora citrophthora Phytophthora cryptogea Phytophthora drechsleri Phytophthora erythroseptica Phytophthora europaea Phytophthora fragariae var fragariae Phytophthora fragariae var rubi Phytophthora glovera Phytophthora gonapodyides Phytophthora heveae Phytophthora hibernalis Phytophthora infestans Phytophthora kernoviae Phytophthora lagoariana -

Phytophthora Fragariae: Phytophthora Fragariae Var. Fragariae
23/3/2018 All data for a single taxon ***Tell us why you value the fungal databases*** Data for Phytophthora fragariae Phytophthora fragariae var. fragariae Hickman 1940 (Oomycetes, Pythiales) ≡Phytophthora fragariae C.J. Hickman 1940 Note: See type variety. Distribution: Asia, Australia, New Zealand, Europe, North America (Canada, USA). Substrate: Roots. Disease Note: Red stele or red core root rot. The most important fungal pathogen of strawberries (Ho & Jong 1988). Foreign races are listed by APHIS as a regulated plant pest. Host: Fragaria x ananassa and Rubus ursinus var. longanobaccus (Rosaceae) are the only known hosts under natural conditions. Infection of other Rosaceae, Amaranthaceae, and Solanaceae can occur via artificial inoculation (Erwin 1996). Supporting Literature: Erwin, D.C., and Ribeiro, O.K. 1996. Phytophthora Diseases Worldwide. APS Press, St. Paul, Minnesota, 562 pages. Ho, H.H., and Jong, S.C. 1988. Phytophthora fragariae. Mycotaxon 31: 305-322. Wilcox, W.F. 1989. Identity, virulence, and isolation frequency of seven Phytophthora spp. causing root rot of raspberry in New York. Phytopathology 79(1): 93-101. Yang, X., Tyler, B.M., and Hong, C. 2017. An expanded phylogeny for the genus Phytophthora. IMA Fungus 8(2): 355-384. Updated on Mar 07, 2018 Fungus-Host - 124 records were found using the criteria: name = Phytophthora fragariae and its synonyms Phytophthora fragariae: Fragaria chiloensis (Red stele.): California - 3250,Canada - 8376,Connecticut - 1431,Oregon - 44, 3250,Washington - 44, 3250, Fragaria elatior Bulgaria -

Immunological Diagnostic of Phytophthora Infestans from Host Tissues (Potato) by ELISA Method
Türk Tarım ve Doğa Bilimleri Dergisi 4(4): 385–392, 2017 TÜRK TURKISH TARIM ve DOĞA BİLİMLERİ JOURNAL of AGRICULTURAL DERGİSİ and NATURAL SCIENCES www.dergipark.gov.tr/turkjans Immunological Diagnostic of Phytophthora infestans from Host Tissues (Potato) by ELISA Method 1Touseef HUSSAIN*, 2Bir Pal SINGH, 3Firoz ANWAR 1Dept. of Life Science, Uttarakhand Technical University, Dehradun, 248009, U.K, India 2ICAR-Central Potato Research Institute, Shimla-171001, H.P, India 3Faculty of Science, King Abdulaziz University, Jeddah, Kingdom of Saudi Arabia-80200 *Corresponding author: [email protected] Received: 06.02.2017 Received in Revised: 11.08.2017 Accepted: 11.08.2017 Abstract Oomycete pathogens of the genus Phytophthora are the most destructive plant pathogens known. They spread mainly through the movement of infested soil, water and infected plants and plant material. Especially damaging as a source of inoculum are those plants/seeds that are infected but do not show signs of symptoms either because the disease has not yet progressed to the stage where symptoms are evident, or due to suppression of symptom development by the use of fungicides. In our indirect ELISA method, P.infestans exhibited strong positive reaction with sporangia (2.256), mycelium (1.256) as well as oospores (2.286) whereas no reaction with other fungal pathogens of potato. P.infestans was detected by indirect ELISA in potato leaves (1.212) and tubers (1.201). Our study was to confirm the detection of P.infestans irrespective of inoculum present in the host tissues (0.435), by which planting material by quarantine dept., horticultural dept. from one state to another. -

Molecular Diagnostics and Detection of Oomycetes on Fiber Crops
plants Review Molecular Diagnostics and Detection of Oomycetes on Fiber Crops Tuhong Wang 1 , Chunsheng Gao 1, Yi Cheng 1 , Zhimin Li 1, Jia Chen 1, Litao Guo 1 and Jianping Xu 1,2,* 1 Institute of Bast Fiber Crops and Center of Southern Economic Crops, Chinese Academy of Agricultural Sciences, Changsha 410205, China; [email protected] (T.W.); [email protected] (C.G.); [email protected] (Y.C.); [email protected] (Z.L.); [email protected] (J.C.); [email protected] (L.G.) 2 Department of Biology, McMaster University, Hamilton, ON L8S 4K1, Canada * Correspondence: [email protected] Received: 15 May 2020; Accepted: 15 June 2020; Published: 19 June 2020 Abstract: Fiber crops are an important group of economic plants. Traditionally cultivated for fiber, fiber crops have also become sources of other materials such as food, animal feed, cosmetics and medicine. Asia and America are the two main production areas of fiber crops in the world. However, oomycete diseases have become an important factor limiting their yield and quality, causing devastating consequences for the production of fiber crops in many regions. To effectively control oomycete pathogens and reduce their negative impacts on these crops, it is very important to have fast and accurate detection systems, especially in the early stages of infection. With the rapid development of molecular biology, the diagnosis of plant pathogens has progressed from relying on traditional morphological features to the increasing use of molecular methods. The objective of this paper was to review the current status of research on molecular diagnosis of oomycete pathogens on fiber crops.