Metabolic Evaluation of Epilepsy: a Diagnostic Algorithm with Focus on Treatable Conditions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Regulation of Skeletal Muscle Glucose Transport and Glucose Metabolism by Exercise Training
nutrients Review Regulation of Skeletal Muscle Glucose Transport and Glucose Metabolism by Exercise Training Parker L. Evans 1,2,3, Shawna L. McMillin 1,2,3 , Luke A. Weyrauch 1,2,3 and Carol A. Witczak 1,2,3,4,* 1 Department of Kinesiology, East Carolina University, Greenville, NC 27858, USA; [email protected] (P.L.E.); [email protected] (S.L.M.); [email protected] (L.A.W.) 2 Department of Physiology, Brody School of Medicine, East Carolina University, Greenville, NC 27834, USA 3 East Carolina Diabetes & Obesity Institute, East Carolina University, Greenville, NC 27834, USA 4 Department of Biochemistry & Molecular Biology, Brody School of Medicine, East Carolina University, Greenville, NC 27834, USA * Correspondence: [email protected]; Tel.: +1-252-744-1224 Received: 8 September 2019; Accepted: 8 October 2019; Published: 12 October 2019 Abstract: Aerobic exercise training and resistance exercise training are both well-known for their ability to improve human health; especially in individuals with type 2 diabetes. However, there are critical differences between these two main forms of exercise training and the adaptations that they induce in the body that may account for their beneficial effects. This article reviews the literature and highlights key gaps in our current understanding of the effects of aerobic and resistance exercise training on the regulation of systemic glucose homeostasis, skeletal muscle glucose transport and skeletal muscle glucose metabolism. Keywords: aerobic exercise; blood glucose; functional overload; GLUT; hexokinase; insulin resistance; resistance exercise; SGLT; type 2 diabetes; weightlifting 1. Introduction Exercise training is defined as planned bouts of physical activity which repeatedly occur over a duration of time lasting from weeks to years. -

Genes in Eyecare Geneseyedoc 3 W.M
Genes in Eyecare geneseyedoc 3 W.M. Lyle and T.D. Williams 15 Mar 04 This information has been gathered from several sources; however, the principal source is V. A. McKusick’s Mendelian Inheritance in Man on CD-ROM. Baltimore, Johns Hopkins University Press, 1998. Other sources include McKusick’s, Mendelian Inheritance in Man. Catalogs of Human Genes and Genetic Disorders. Baltimore. Johns Hopkins University Press 1998 (12th edition). http://www.ncbi.nlm.nih.gov/Omim See also S.P.Daiger, L.S. Sullivan, and B.J.F. Rossiter Ret Net http://www.sph.uth.tmc.edu/Retnet disease.htm/. Also E.I. Traboulsi’s, Genetic Diseases of the Eye, New York, Oxford University Press, 1998. And Genetics in Primary Eyecare and Clinical Medicine by M.R. Seashore and R.S.Wappner, Appleton and Lange 1996. M. Ridley’s book Genome published in 2000 by Perennial provides additional information. Ridley estimates that we have 60,000 to 80,000 genes. See also R.M. Henig’s book The Monk in the Garden: The Lost and Found Genius of Gregor Mendel, published by Houghton Mifflin in 2001 which tells about the Father of Genetics. The 3rd edition of F. H. Roy’s book Ocular Syndromes and Systemic Diseases published by Lippincott Williams & Wilkins in 2002 facilitates differential diagnosis. Additional information is provided in D. Pavan-Langston’s Manual of Ocular Diagnosis and Therapy (5th edition) published by Lippincott Williams & Wilkins in 2002. M.A. Foote wrote Basic Human Genetics for Medical Writers in the AMWA Journal 2002;17:7-17. A compilation such as this might suggest that one gene = one disease. -

Sialic Acid Storage Disease
Sialic acid storage disease Description Sialic acid storage disease is an inherited disorder that primarily affects the nervous system. People with sialic acid storage disease have signs and symptoms that may vary widely in severity. This disorder is generally classified into one of three forms: infantile free sialic acid storage disease, Salla disease, and intermediate severe Salla disease. Infantile free sialic acid storage disease (ISSD) is the most severe form of this disorder. Babies with this condition have severe developmental delay, weak muscle tone ( hypotonia), and failure to gain weight and grow at the expected rate (failure to thrive). They may have unusual facial features that are often described as "coarse," seizures, bone malformations, an enlarged liver and spleen (hepatosplenomegaly), and an enlarged heart (cardiomegaly). The abdomen may be swollen due to the enlarged organs and an abnormal buildup of fluid in the abdominal cavity (ascites). Affected infants may have a condition called hydrops fetalis in which excess fluid accumulates in the body before birth. Children with this severe form of the condition usually live only into early childhood. Salla disease is a less severe form of sialic acid storage disease. Babies with Salla disease usually begin exhibiting hypotonia during the first year of life and go on to experience progressive neurological problems. Signs and symptoms of Salla disease include intellectual disability and developmental delay, seizures, problems with movement and balance (ataxia), abnormal tensing of the muscles (spasticity), and involuntary slow, sinuous movements of the limbs (athetosis). Individuals with Salla disease usually survive into adulthood. People with intermediate severe Salla disease have signs and symptoms that fall between those of ISSD and Salla disease in severity. -

WES Gene Package Multiple Congenital Anomalie.Xlsx
Whole Exome Sequencing Gene package Multiple congenital anomalie, version 5, 1‐2‐2018 Technical information DNA was enriched using Agilent SureSelect Clinical Research Exome V2 capture and paired‐end sequenced on the Illumina platform (outsourced). The aim is to obtain 8.1 Giga base pairs per exome with a mapped fraction of 0.99. The average coverage of the exome is ~50x. Duplicate reads are excluded. Data are demultiplexed with bcl2fastq Conversion Software from Illumina. Reads are mapped to the genome using the BWA‐MEM algorithm (reference: http://bio‐bwa.sourceforge.net/). Variant detection is performed by the Genome Analysis Toolkit HaplotypeCaller (reference: http://www.broadinstitute.org/gatk/). The detected variants are filtered and annotated with Cartagenia software and classified with Alamut Visual. It is not excluded that pathogenic mutations are being missed using this technology. At this moment, there is not enough information about the sensitivity of this technique with respect to the detection of deletions and duplications of more than 5 nucleotides and of somatic mosaic mutations (all types of sequence changes). HGNC approved Phenotype description including OMIM phenotype ID(s) OMIM median depth % covered % covered % covered gene symbol gene ID >10x >20x >30x A4GALT [Blood group, P1Pk system, P(2) phenotype], 111400 607922 101 100 100 99 [Blood group, P1Pk system, p phenotype], 111400 NOR polyagglutination syndrome, 111400 AAAS Achalasia‐addisonianism‐alacrimia syndrome, 231550 605378 73 100 100 100 AAGAB Keratoderma, palmoplantar, -

The Genetic Landscape of the Human Solute Carrier (SLC) Transporter Superfamily
Human Genetics (2019) 138:1359–1377 https://doi.org/10.1007/s00439-019-02081-x ORIGINAL INVESTIGATION The genetic landscape of the human solute carrier (SLC) transporter superfamily Lena Schaller1 · Volker M. Lauschke1 Received: 4 August 2019 / Accepted: 26 October 2019 / Published online: 2 November 2019 © The Author(s) 2019 Abstract The human solute carrier (SLC) superfamily of transporters is comprised of over 400 membrane-bound proteins, and plays essential roles in a multitude of physiological and pharmacological processes. In addition, perturbation of SLC transporter function underlies numerous human diseases, which renders SLC transporters attractive drug targets. Common genetic polymorphisms in SLC genes have been associated with inter-individual diferences in drug efcacy and toxicity. However, despite their tremendous clinical relevance, epidemiological data of these variants are mostly derived from heterogeneous cohorts of small sample size and the genetic SLC landscape beyond these common variants has not been comprehensively assessed. In this study, we analyzed Next-Generation Sequencing data from 141,456 individuals from seven major human populations to evaluate genetic variability, its functional consequences, and ethnogeographic patterns across the entire SLC superfamily of transporters. Importantly, of the 204,287 exonic single-nucleotide variants (SNVs) which we identifed, 99.8% were present in less than 1% of analyzed alleles. Comprehensive computational analyses using 13 partially orthogonal algorithms that predict the functional impact of genetic variations based on sequence information, evolutionary conserva- tion, structural considerations, and functional genomics data revealed that each individual genome harbors 29.7 variants with putative functional efects, of which rare variants account for 18%. Inter-ethnic variability was found to be extensive, and 83% of deleterious SLC variants were only identifed in a single population. -

Centometabolic® COMBINING GENETIC and BIOCHEMICAL TESTING for the FAST and COMPREHENSIVE DIAGNOSTIC of METABOLIC DISORDERS
CentoMetabolic® COMBINING GENETIC AND BIOCHEMICAL TESTING FOR THE FAST AND COMPREHENSIVE DIAGNOSTIC OF METABOLIC DISORDERS GENE ASSOCIATED DISEASE(S) ABCA1 HDL deficiency, familial, 1; Tangier disease ABCB4 Cholestasis, progressive familial intrahepatic 3 ABCC2 Dubin-Johnson syndrome ABCD1 Adrenoleukodystrophy; Adrenomyeloneuropathy, adult ABCD4 Methylmalonic aciduria and homocystinuria, cblJ type ABCG5 Sitosterolemia 2 ABCG8 Sitosterolemia 1 ACAT1 Alpha-methylacetoacetic aciduria ADA Adenosine deaminase deficiency AGA Aspartylglucosaminuria AGL Glycogen storage disease IIIa; Glycogen storage disease IIIb AGPS Rhizomelic chondrodysplasia punctata, type 3 AGXT Hyperoxaluria, primary, type 1 ALAD Porphyria, acute hepatic ALAS2 Anemia, sideroblastic, 1; Protoporphyria, erythropoietic, X-linked ALDH4A1 Hyperprolinemia, type II ALDOA Glycogen storage disease XII ALDOB Fructose intolerance, hereditary ALG3 Congenital disorder of glycosylation, type Id ALPL Hypophosphatasia, adult; Hypophosphatasia, childhood; Hypophosphatasia, infantile; Odontohypophosphatasia ANTXR2 Hyaline fibromatosis syndrome APOA2 Hypercholesterolemia, familial, modifier of APOA5 Hyperchylomicronemia, late-onset APOB Hypercholesterolemia, familial, 2 APOC2 Hyperlipoproteinemia, type Ib APOE Sea-blue histiocyte disease; Hyperlipoproteinemia, type III ARG1 Argininemia ARSA Metachromatic leukodystrophy ARSB Mucopolysaccharidosis type VI (Maroteaux-Lamy) 1 GENE ASSOCIATED DISEASE(S) ASAH1 Farber lipogranulomatosis; Spinal muscular atrophy with progressive myoclonic epilepsy -

Table S1. Disease Classification and Disease-Reaction Association
Table S1. Disease classification and disease-reaction association Disorder class Associated reactions cross Disease Ref[Goh check et al. -

The Genetic Relationship Between Paroxysmal Movement Disorders and Epilepsy
Review article pISSN 2635-909X • eISSN 2635-9103 Ann Child Neurol 2020;28(3):76-87 https://doi.org/10.26815/acn.2020.00073 The Genetic Relationship between Paroxysmal Movement Disorders and Epilepsy Hyunji Ahn, MD, Tae-Sung Ko, MD Department of Pediatrics, Asan Medical Center Children’s Hospital, University of Ulsan College of Medicine, Seoul, Korea Received: May 1, 2020 Revised: May 12, 2020 Seizures and movement disorders both involve abnormal movements and are often difficult to Accepted: May 24, 2020 distinguish due to their overlapping phenomenology and possible etiological commonalities. Par- oxysmal movement disorders, which include three paroxysmal dyskinesia syndromes (paroxysmal Corresponding author: kinesigenic dyskinesia, paroxysmal non-kinesigenic dyskinesia, paroxysmal exercise-induced dys- Tae-Sung Ko, MD kinesia), hemiplegic migraine, and episodic ataxia, are important examples of conditions where Department of Pediatrics, Asan movement disorders and seizures overlap. Recently, many articles describing genes associated Medical Center Children’s Hospital, University of Ulsan College of with paroxysmal movement disorders and epilepsy have been published, providing much infor- Medicine, 88 Olympic-ro 43-gil, mation about their molecular pathology. In this review, we summarize the main genetic disorders Songpa-gu, Seoul 05505, Korea that results in co-occurrence of epilepsy and paroxysmal movement disorders, with a presenta- Tel: +82-2-3010-3390 tion of their genetic characteristics, suspected pathogenic mechanisms, and detailed descriptions Fax: +82-2-473-3725 of paroxysmal movement disorders and seizure types. E-mail: [email protected] Keywords: Dyskinesias; Movement disorders; Seizures; Epilepsy Introduction ies, and paroxysmal dyskinesias [3,4]. Paroxysmal dyskinesias are an important disease paradigm asso- Movement disorders often arise from the basal ganglia nuclei or ciated with overlapping movement disorders and seizures [5]. -

Antenatal Diagnosis of Inborn Errors Ofmetabolism
816 ArchivesofDiseaseinChildhood 1991;66: 816-822 CURRENT PRACTICE Arch Dis Child: first published as 10.1136/adc.66.7_Spec_No.816 on 1 July 1991. Downloaded from Antenatal diagnosis of inborn errors of metabolism M A Cleary, J E Wraith The introduction of experimental treatment for Sample requirement and techniques used in lysosomal storage disorders and the increasing prenatal diagnosis understanding of the molecular defects behind By far the majority of antenatal diagnoses are many inborn errors have overshadowed the fact performed on samples obtained by either that for many affected families the best that can amniocentesis or chorion villus biopsy. For be offered is a rapid, accurate prenatal diag- some disorders, however, the defect is not nostic service. Many conditions remain at best detectable in this material and more invasive only partially treatable and as a consequence the methods have been applied to obtain a diagnos- majority of parents seek antenatal diagnosis in tic sample. subsequent pregnancies, particularly for those disorders resulting in a poor prognosis in terms of either life expectancy or normal neurological FETAL LIVER BIOPSY development. Fetal liver biopsy has been performed to The majority of inborn errors result from a diagnose ornithine carbamoyl transferase defi- specific enzyme deficiency, but in some the ciency and primary hyperoxaluria type 1. primary defect is in a transport system or Glucose-6-phosphatase deficiency (glycogen enzyme cofactor. In some conditions the storage disease type I) could also be detected by biochemical defect is limited to specific tissues this method. The technique, however, is inva- only and this serves to restrict the material avail- sive and can be performed by only a few highly able for antenatal diagnosis for these disorders. -

Transporters
Alexander, S. P. H., Kelly, E., Mathie, A., Peters, J. A., Veale, E. L., Armstrong, J. F., Faccenda, E., Harding, S. D., Pawson, A. J., Sharman, J. L., Southan, C., Davies, J. A., & CGTP Collaborators (2019). The Concise Guide to Pharmacology 2019/20: Transporters. British Journal of Pharmacology, 176(S1), S397-S493. https://doi.org/10.1111/bph.14753 Publisher's PDF, also known as Version of record License (if available): CC BY Link to published version (if available): 10.1111/bph.14753 Link to publication record in Explore Bristol Research PDF-document This is the final published version of the article (version of record). It first appeared online via Wiley at https://bpspubs.onlinelibrary.wiley.com/doi/full/10.1111/bph.14753. Please refer to any applicable terms of use of the publisher. University of Bristol - Explore Bristol Research General rights This document is made available in accordance with publisher policies. Please cite only the published version using the reference above. Full terms of use are available: http://www.bristol.ac.uk/red/research-policy/pure/user-guides/ebr-terms/ S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology (2019) 176, S397–S493 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters Stephen PH Alexander1 , Eamonn Kelly2, Alistair Mathie3 ,JohnAPeters4 , Emma L Veale3 , Jane F Armstrong5 , Elena Faccenda5 ,SimonDHarding5 ,AdamJPawson5 , Joanna L Sharman5 , Christopher Southan5 , Jamie A Davies5 and CGTP Collaborators 1School of Life Sciences, -

Downloaded from the App Store and Nucleobase, Nucleotide and Nucleic Acid Metabolism 7 Google Play
Hoytema van Konijnenburg et al. Orphanet J Rare Dis (2021) 16:170 https://doi.org/10.1186/s13023-021-01727-2 REVIEW Open Access Treatable inherited metabolic disorders causing intellectual disability: 2021 review and digital app Eva M. M. Hoytema van Konijnenburg1†, Saskia B. Wortmann2,3,4†, Marina J. Koelewijn2, Laura A. Tseng1,4, Roderick Houben6, Sylvia Stöckler‑Ipsiroglu5, Carlos R. Ferreira7 and Clara D. M. van Karnebeek1,2,4,8* Abstract Background: The Treatable ID App was created in 2012 as digital tool to improve early recognition and intervention for treatable inherited metabolic disorders (IMDs) presenting with global developmental delay and intellectual disabil‑ ity (collectively ‘treatable IDs’). Our aim is to update the 2012 review on treatable IDs and App to capture the advances made in the identifcation of new IMDs along with increased pathophysiological insights catalyzing therapeutic development and implementation. Methods: Two independent reviewers queried PubMed, OMIM and Orphanet databases to reassess all previously included disorders and therapies and to identify all reports on Treatable IDs published between 2012 and 2021. These were included if listed in the International Classifcation of IMDs (ICIMD) and presenting with ID as a major feature, and if published evidence for a therapeutic intervention improving ID primary and/or secondary outcomes is avail‑ able. Data on clinical symptoms, diagnostic testing, treatment strategies, efects on outcomes, and evidence levels were extracted and evaluated by the reviewers and external experts. The generated knowledge was translated into a diagnostic algorithm and updated version of the App with novel features. Results: Our review identifed 116 treatable IDs (139 genes), of which 44 newly identifed, belonging to 17 ICIMD categories. -
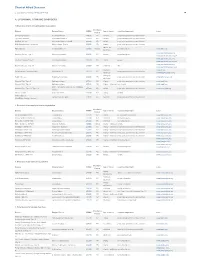
Pdf NTSAD Chart of Allied Diseases
Chart of Allied Diseases Last Updated: Monday, 19 May 2014 17:02 A. LYSOSOMAL STORAGE DISORDERS 1) Disorders of lipid and sphingolipid degradation Inheritance Disease Enzyme Defect OMIM# Age of Onset Cognitive Impairment Links Pattern GM1 Gangliosidosis b-Galactosidase-1 230500 AR variable progressive psychomotor deterioration Tay-Sachs Disease b-Hexosaminidase A 272800 AR variable progressive psychomotor deterioration Sandhoff Disease b-Hexosaminidases A and B 268800 AR variable progressive psychomotor deterioration GM2 Gangliodisosis, AB variant GM2 Activator Protein 272750 AR infancy progressive psychomotor deterioration adolesence - Fabry Disease 8-Galactosidase A 301500 X-linked normal intelligence www.fabry.org adulthood www.gaucherdisease.org, Gaucher Disease, Type 1 Glucocerebrosidase 230800 AR variable normal intelligence www.gaucherdisease.org.uk www.gaucherdisease.org, Gaucher Disease, Type II Glucocerebrosidase 230900 AR infancy severe www.gaucherdisease.org.uk www.gaucherdisease.org, Gaucher Disease, Type III Glucocerebrosidase 231000 AR childhood mild www.gaucherdisease.org.uk infancy to www.ulf.org, Metachromatic Leukodystrophy Arylsulfatase A 250100 AR progressive psychomotor deterioration adulthood www.MLDFoundation.org infancy to Krabbe Disease Galactosylceramidase 245200 AR progressive psychomotor deterioration www.huntershope.org adulthood Niemann-Pick, Type A Sphingomyelinase 257200 AR infancy progressive psychomotor deterioration www.nnpdf.org Niemann-Pick, Type B Sphingomyelinase 607616 AR infancy - childhood none