Males Mosaic for Mutations in the X-Linked EFNB1 Gene Are More Severely Affected Than True Hemizygotes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
![Downloaded from [266]](https://docslib.b-cdn.net/cover/7352/downloaded-from-266-347352.webp)
Downloaded from [266]
Patterns of DNA methylation on the human X chromosome and use in analyzing X-chromosome inactivation by Allison Marie Cotton B.Sc., The University of Guelph, 2005 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in The Faculty of Graduate Studies (Medical Genetics) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) January 2012 © Allison Marie Cotton, 2012 Abstract The process of X-chromosome inactivation achieves dosage compensation between mammalian males and females. In females one X chromosome is transcriptionally silenced through a variety of epigenetic modifications including DNA methylation. Most X-linked genes are subject to X-chromosome inactivation and only expressed from the active X chromosome. On the inactive X chromosome, the CpG island promoters of genes subject to X-chromosome inactivation are methylated in their promoter regions, while genes which escape from X- chromosome inactivation have unmethylated CpG island promoters on both the active and inactive X chromosomes. The first objective of this thesis was to determine if the DNA methylation of CpG island promoters could be used to accurately predict X chromosome inactivation status. The second objective was to use DNA methylation to predict X-chromosome inactivation status in a variety of tissues. A comparison of blood, muscle, kidney and neural tissues revealed tissue-specific X-chromosome inactivation, in which 12% of genes escaped from X-chromosome inactivation in some, but not all, tissues. X-linked DNA methylation analysis of placental tissues predicted four times higher escape from X-chromosome inactivation than in any other tissue. Despite the hypomethylation of repetitive elements on both the X chromosome and the autosomes, no changes were detected in the frequency or intensity of placental Cot-1 holes. -

Contributions of the Renin Angiotensin System to Fear Memory and Fear Conditioned Cardiovascular Responses
Contributions of the Renin Angiotensin System to Fear Memory and Fear Conditioned Cardiovascular Responses by Adam Swiercz B.S. in Biology, May 2006, The George Washington University M.P.S. in Molecular Biotechnology, May 2009, The George Washington University M.S. in Physiology, May 2011, Georgetown University A Dissertation submitted to The Faculty of The Columbian College of Arts & Sciences of The George Washington University in partial fulfillment of the requirements for the degree of Doctor of Philosophy January 10, 2020 Dissertation co-directed by Paul J. Marvar Associate Professor of Pharmacology and Physiology and David Mendelowitz Professor of Pharmacology & Physiology The Columbian College of Arts and Sciences of The George Washington University certifies that Adam Swiercz has passed the Final Examination for the degree of Doctor of Philosophy as of October 2nd, 2019. This is the final and approved form of the dissertation. Contributions of the Renin Angiotensin System to Fear Memory and Fear Conditioned Cardiovascular Responses Adam Swiercz Dissertation Research Committee: Paul J. Marvar, Associate Professor of Pharmacology & Physiology, Dissertation Co-Director David Mendelowitz, Professor of Pharmacology & Physiology, Dissertation Co-Director Abigail Polter, Assistant Professor of Pharmacology & Physiology, Committee Member Colin Young, Assistant Professor of Pharmacology & Physiology, Committee Member ii © Copyright 2020 by Adam Swiercz All rights reserved iii Acknowledgements I would like to thank and acknowledge Dr. Paul Marvar, whose mentorship has made this dissertation possible. It has been a pleasure working in your lab, and I am truly grateful for your support and encouragement throughout the years. Thanks to the current and former members of the Marvar lab who have made my time at GW a rewarding and enjoyable experience. -

Oncogenic Inhibition by a Deleted in Liver Cancer Gene Requires Cooperation Between Tensin Binding and Rho-Specific Gtpase-Activating Protein Activities
Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities Xiaolan Qian*, Guorong Li*, Holly K. Asmussen*, Laura Asnaghi*, William C. Vass*, Richard Braverman*, Kenneth M. Yamada†, Nicholas C. Popescu‡, Alex G. Papageorge*, and Douglas R. Lowy*§ *Laboratory of Cellular Oncology and ‡Laboratory of Experimental Carcinogenesis, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892; and †Laboratory of Cell and Developmental Biology, National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD 20892 Communicated by Ira Pastan, National Institutes of Health, Bethesda, MD, April 2, 2007 (received for review March 1, 2007) The three deleted in liver cancer genes (DLC1–3) encode Rho- The prototypic member, designated DLC1, is localized to GTPase-activating proteins (RhoGAPs) whose expression is fre- chromosome 8p21–22 in a region that is commonly deleted in quently down-regulated or silenced in a variety of human malig- hepatocellular carcinoma (5). Its expression is frequently down- nancies. The RhoGAP activity is required for full DLC-dependent regulated or silenced in various solid tumors and hematologic tumor suppressor activity. Here we report that DLC1 and DLC3 bind malignancies, predominantly by promoter methylation (6–13). to human tensin1 and its chicken homolog. The binding has been Ectopic reexpression in DLC1-deficient cancer cell lines can mapped to the tensin Src homology 2 (SH2) and phosphotyrosine suppress cell proliferation, induce apoptosis, and reduce tumor- binding (PTB) domains at the C terminus of tensin proteins. Distinct igenicity. The RhoGAP activity appears to be required for these DLC1 sequences are required for SH2 and PTB binding. -

WO 2012/174282 A2 20 December 2012 (20.12.2012) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2012/174282 A2 20 December 2012 (20.12.2012) P O P C T (51) International Patent Classification: David [US/US]; 13539 N . 95th Way, Scottsdale, AZ C12Q 1/68 (2006.01) 85260 (US). (21) International Application Number: (74) Agent: AKHAVAN, Ramin; Caris Science, Inc., 6655 N . PCT/US20 12/0425 19 Macarthur Blvd., Irving, TX 75039 (US). (22) International Filing Date: (81) Designated States (unless otherwise indicated, for every 14 June 2012 (14.06.2012) kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, English (25) Filing Language: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, Publication Language: English DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, KR, (30) Priority Data: KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, 61/497,895 16 June 201 1 (16.06.201 1) US MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, 61/499,138 20 June 201 1 (20.06.201 1) US OM, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SC, SD, 61/501,680 27 June 201 1 (27.06.201 1) u s SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, 61/506,019 8 July 201 1(08.07.201 1) u s TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. -

Egfr Activates a Taz-Driven Oncogenic Program in Glioblastoma
EGFR ACTIVATES A TAZ-DRIVEN ONCOGENIC PROGRAM IN GLIOBLASTOMA by Minling Gao A thesis submitted to Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland March 2020 ©2020 Minling Gao All rights reserved Abstract Hyperactivated EGFR signaling is associated with about 45% of Glioblastoma (GBM), the most aggressive and lethal primary brain tumor in humans. However, the oncogenic transcriptional events driven by EGFR are still incompletely understood. We studied the role of the transcription factor TAZ to better understand master transcriptional regulators in mediating the EGFR signaling pathway in GBM. The transcriptional coactivator with PDZ- binding motif (TAZ) and its paralog gene, the Yes-associated protein (YAP) are two transcriptional co-activators that play important roles in multiple cancer types and are regulated in a context-dependent manner by various upstream signaling pathways, e.g. the Hippo, WNT and GPCR signaling. In GBM cells, TAZ functions as an oncogene that drives mesenchymal transition and radioresistance. This thesis intends to broaden our understanding of EGFR signaling and TAZ regulation in GBM. In patient-derived GBM cell models, EGF induced TAZ and its known gene targets through EGFR and downstream tyrosine kinases (ERK1/2 and STAT3). In GBM cells with EGFRvIII, an EGF-independent and constitutively active mutation, TAZ showed EGF- independent hyperactivation when compared to EGFRvIII-negative cells. These results revealed a novel EGFR-TAZ signaling axis in GBM cells. The second contribution of this thesis is that we performed next-generation sequencing to establish the first genome-wide map of EGF-induced TAZ target genes. -

Differentially Expressed Genes in Aneurysm Tissue Compared With
On-line Table: Differentially expressed genes in aneurysm tissue compared with those in control tissue Fold False Discovery Direction of Gene Entrez Gene Name Function Change P Value Rate (q Value) Expression AADAC Arylacetamide deacetylase Positive regulation of triglyceride 4.46 1.33E-05 2.60E-04 Up-regulated catabolic process ABCA6 ATP-binding cassette, subfamily A (ABC1), Integral component of membrane 3.79 9.15E-14 8.88E-12 Up-regulated member 6 ABCC3 ATP-binding cassette, subfamily C (CFTR/MRP), ATPase activity, coupled to 6.63 1.21E-10 7.33E-09 Up-regulated member 3 transmembrane movement of substances ABI3 ABI family, member 3 Peptidyl-tyrosine phosphorylation 6.47 2.47E-05 4.56E-04 Up-regulated ACKR1 Atypical chemokine receptor 1 (Duffy blood G-protein–coupled receptor signaling 3.80 7.95E-10 4.18E-08 Up-regulated group) pathway ACKR2 Atypical chemokine receptor 2 G-protein–coupled receptor signaling 0.42 3.29E-04 4.41E-03 Down-regulated pathway ACSM1 Acyl-CoA synthetase medium-chain family Energy derivation by oxidation of 9.87 1.70E-08 6.52E-07 Up-regulated member 1 organic compounds ACTC1 Actin, ␣, cardiac muscle 1 Negative regulation of apoptotic 0.30 7.96E-06 1.65E-04 Down-regulated process ACTG2 Actin, ␥2, smooth muscle, enteric Blood microparticle 0.29 1.61E-16 2.36E-14 Down-regulated ADAM33 ADAM domain 33 Integral component of membrane 0.23 9.74E-09 3.95E-07 Down-regulated ADAM8 ADAM domain 8 Positive regulation of tumor necrosis 4.69 2.93E-04 4.01E-03 Up-regulated factor (ligand) superfamily member 11 production ADAMTS18 -

Single Cell Analysis Reveals X Chromosome Upregulation Is Not
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.18.452817; this version posted July 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Single cell analysis reveals X chromosome upregulation is not global and 2 primarily belongs to ancestral genes in pre-gastrulation embryos 3 Naik C H, Chandel D, and Gayen S* 4 Department of Molecular Reproduction, Development and Genetics, Indian Institute of Science, 5 Bangalore-560012, India. 6 *Correspondence: [email protected] 7 8 Abstract 9 Recent studies have provided substantial evidence supporting Ohno's hypothesis that 10 upregulation of active X chromosome genes balances the dosage of X-linked gene expression 11 relative to autosomal genes. However, the dynamics of X-chromosome upregulation (XCU) 12 during early development remain poorly Pre-gastrulation embryos E6.50 13 understood. Here, we have profiled the E6.25 E5.5 14 dynamics of XCU in different lineages of 15 female pre-gastrulation mouse embryos at 16 single cell level through allele-specific single 17 cell RNA-seq analysis. We found dynamic 18 XCU upon initiation of random X- Epiblast cells 19 chromosome inactivation (XCI) in epiblast Onset of Random X-inactivation 20 cells and cells of extraembryonic lineages, 21 which undergo imprinted XCI, also harbored 22 upregulated active-X chromosome. On the 23 other hand, the extent of XCU remains 24 controversial till date. -
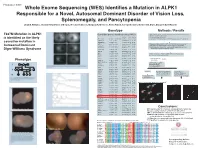
T237M Mutation in ALPK1 Is Identified As the Likely Causative Mutation In
Program #: 3367 Whole Exome Sequencing (WES) Identifies a Mutation in ALPK1 Responsible for a Novel, Autosomal Dominant Disorder of Vision Loss, Splenomegaly, and Pancytopenia Lloyd B. Williams, Chad D. Huff, Denise J. Morgan, Rosann Robinson, Margaux A. Morrison, Krista Kinard, George Rodgers, Kathleen B. Digre, Margaret M. DeAngelis Genotype Methods / Results WES on 4 individuals - 2 affected II-3 III-2, and 2 unaffected II-2 and III-3 T237M Mutation in ALPK1 Candi date genes identified using VAAST Illuimina Truseq Enrichment Kit position Allele Protein Quantitative PCR enrichment is identified as the likely Gene name p-value chromosome (hg19) change change Sequencing with Illumina HiSeq 2000 101 cycle paired end sequencing PRAMEF11 6.10E-06 chr1 12887174 C->T R->H causative mutation in ANKRD20A4 7.32E-06 chr9 69423637 G->A E->K MRPL4 8.55E-06 chr19 10367459 C->T R->W Mapped and aligned with Picard Tools (http://picard.sourceforge.net) FAM90A10 9.77E-06 chr8 7629232 G->T A->S SNPs and indels identified with Genome analysis toolkit (GATK) Autosomal Dominant Manual inspection and curation was done with Intergrative Genomics GOLGA6L10 9.77E-06 chr15 82635194 T->C E->G Viewer (http://www.broadinstitute.org/igv) Digre-Williams Syndrome FAM90A20 1.28E-05 chr8 7155458 C->G A->G EEF1A1 1.65E-05 chr6 74228474 C->T R->H MSI1 1.65E-05 chr12 120800875 C->T V->M VAAST - compares to HGMD, dbSNP, ESP, and1000 Genomes VDAC2 1.71E-05 chr10 76980685 G->T A->S panels to rule out common variants. PIM1 2.08E-05 chr6 37138779 A->T K->M USP11 2.26E-05 chrX -

Estrogen's Impact on the Specialized Transcriptome, Brain, and Vocal
Estrogen’s Impact on the Specialized Transcriptome, Brain, and Vocal Learning Behavior of a Sexually Dimorphic Songbird by Ha Na Choe Department of Molecular Genetics & Microbiology Duke University Date:_____________________ Approved: ___________________________ Erich D. Jarvis, Supervisor ___________________________ Hiroaki Matsunami ___________________________ Debra Silver ___________________________ Dong Yan ___________________________ Gregory Crawford Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Molecular Genetics & Microbiology in the Graduate School of Duke University 2020 ABSTRACT Estrogen’s Impact on the Specialized Transcriptome, Brain, and Vocal Learning Behavior of a Sexually Dimorphic Songbird by Ha Na Choe Department of Molecular Genetics & Microbiology Duke University Date:_________________________ Approved: ___________________________ Erich D. Jarvis, Supervisor ___________________________ Hiroaki Matsunami ___________________________ Debra Silver ___________________________ Dong Yan ___________________________ Gregory Crawford Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Molecular Genetics & Microbiology in the Graduate School of Duke University 2020 Copyright by Ha Na Choe 2020 Abstract The song system of the zebra finch (Taeniopygia guttata) is highly sexually dimorphic, where only males develop the neural structures necessary to learn and produce learned vocalizations -

Males Mosaic for Mutations in the X-Linked EFNB1 Gene Are More Severely Affected Than True Hemizygotes
Human Molecular Genetics, 2013, Vol. 22, No. 8 1654–1662 doi:10.1093/hmg/ddt015 Advance Access published on January 17, 2013 Cellular interference in craniofrontonasal syndrome: males mosaic for mutations in the X-linked EFNB1 gene are more severely affected than true hemizygotes Stephen R.F. Twigg1, Christian Babbs1, Marijke E.P. van den Elzen3, Anne Goriely1, Stephen Taylor2, Simon J. McGowan2, Eleni Giannoulatou1,2, Lorne Lonie5, Jiannis Ragoussis5, Elham Sadighi Akha6, Samantha J.L. Knight6, Roseli M. Zechi-Ceide7, Jeannette A.M. Hoogeboom4, Barbara R. Pober8, Helga V. Toriello9, Steven A. Wall10, M. Rita Passos-Bueno11, Han G. Downloaded from Brunner12, Irene M.J. Mathijssen3 and Andrew O.M. Wilkie1,10,∗ 1Clinical Genetics, 2Computational Biology Research Group, Weatherall Institute of Molecular Medicine, University of 3 4 Oxford, Oxford, UK, Department of Plastic and Reconstructive Surgery, Department of Clinical Genetics, Erasmus http://hmg.oxfordjournals.org/ MC, University Medical Center, Rotterdam, The Netherlands, 5The Genomics Group, 6NIHR Biomedical Research Centre Oxford, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK, 7Department of Clinical Genetics, Hospital of Rehabilitation and Craniofacial Anomalies, University of Sa˜o Paulo, Sa˜o Paulo, Brazil, 8Department of Pediatrics, Harvard Medical School and Massachusetts General Hospital, Boston, MA, USA, 9College of Human Medicine, Michigan State University, Grand Rapids, MI, USA, 10Craniofacial Unit, Oxford University Hospitals NHS Trust, Oxford, -

Rapid Isolation of Glomeruli Coupled with Gene Expression Profiling Identifies Downstream Targets in Pod1 Knockout Mice
Rapid Isolation of Glomeruli Coupled with Gene Expression Profiling Identifies Downstream Targets in Pod1 Knockout Mice Shiying Cui,* Chengjin Li,* Masatsugu Ema,† Jordan Weinstein,‡ and Susan E. Quaggin*‡ *The Samuel Lunenfeld Research Institute, University of Toronto, Toronto, Ontario, Canada; †Department of Anatomy and Embryology, Institute of Basic Medical Sciences, Division of Developmental Technology, Laboratory Animal Resource Center, University of Tsukuba, Tsukuba, Ibaraki, Japan; and ‡Division of Nephrology, St. Michael’s Hospital, University of Toronto, Toronto, Ontario, Canada Mouse mutations have provided tremendous insights into the molecular basis of renal and glomerular development. However, genes often play important roles during multiple stages of nephrogenesis, making it difficult to determine the role of a gene in a specific cell lineage such as the podocyte. Conditional gene targeting and chimeric analysis are two possible approaches to dissect the function of genes in specific cell populations. However, these are labor-intensive and costly and require the generation, validation, and analysis of additional transgenic lines. For overcoming these shortcomings and, specifically, for studying the role of gene function in developing glomeruli, a technique to isolate and purify glomeruli from murine embryos was developed. Combined with gene expression profiling, this method was used to identify differentially expressed genes in glomeruli from Pod1 knockout (KO) mice that die in the perinatal period with multiple renal defects. Glomeruli from early developing stages (late S-shape/early capillary loop) onward can be isolated successfully from wild-type and KO kidneys at 18.5 d postcoitus, and RNA can readily be obtained and used for genome-wide microarray analysis. With this approach, 3986 genes that are differently expressed between glomeruli from Pod1 KO and wild-type mice were identified, including a four-fold reduction of ␣ 8 integrin mRNA in glomeruli from Pod1 KO mice that was confirmed by immunostaining. -

Mouse Stard8 Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Stard8 Knockout Project (CRISPR/Cas9) Objective: To create a Stard8 knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Stard8 gene (NCBI Reference Sequence: NM_199018 ; Ensembl: ENSMUSG00000031216 ) is located on Mouse chromosome X. 14 exons are identified, with the ATG start codon in exon 4 and the TGA stop codon in exon 14 (Transcript: ENSMUST00000036606). Exon 4~14 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Exon 4 starts from about 0.03% of the coding region. Exon 4~14 covers 100.0% of the coding region. The size of effective KO region: ~8797 bp. The KO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 3 4 5 6 8 9 10 11 1213 14 Legends Exon of mouse Stard8 Knockout region Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section upstream of start codon is aligned with itself to determine if there are tandem repeats. No significant tandem repeat is found in the dot plot matrix. So this region is suitable for PCR screening or sequencing analysis. Overview of the Dot Plot (down) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section downstream of stop codon is aligned with itself to determine if there are tandem repeats.