Type D Primate Retroviruses: a Review1
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

10.2.2 Nomenclature of Viruses and Taxonomic Groups Bacterial Viruses Such As T1, T2 and Φx174
Classification and nomenclature of viruses History of virus classification • Type of host • Type of disease • Transmition by an arthropod vector • Nucleic acid type • SS or DS • Segmented • Size of the virion • Capsid simmetry • Envelope Nomenclature • Small, icosahedral, single-stranded DNA viruses of animals were called parvoviruses (Latin parvus = small) • Nematode-transmitted polyhedral (icosahedral) viruses of plants were called nepoviruses • Phages T2, T4 and T6 were called T even phages • Serological relationships between viruses were investigated • Distinct strains (serotypes) could be distinguished in serological tests • Antisera against purified virions • Serotypes reflect differences in virus proteins International Committee on Taxonomy of Viruses • Order had to be brought • ICTV was formed in 1966 • Many working groups and is advised by virologists around the world • Rules for the nomenclature and classification of viruses • Considers proposals for new taxonomic groups and virus names • Approved are published in book form and on the web Modern virus classification and nomenclature • Each order, family, subfamily and genus is defined by viral characteristics that are necessary for membership of that group. Classification based on genome sequences • Similarity is represented in diagrams known as phylogenetic trees. • Rooted- the tree begins at a root which is assumed to be the ancestor of the viruses in the tree. • Unrooted- no assumption is made about the ancestor of the viruses in the tree. 10.2.2 Nomenclature of viruses and taxonomic groups Bacterial viruses such as T1, T2 and ϕX174. Viruses of humans and other vertebrates diseases that they cause Examples: measles virus, smallpox virus, foot and mouth disease virus Some viruses city, town or river Examples: Newcastle disease virus, Norwalk virus, Ebola virus Insect viruses Many insect viruses were named after the insect, with an indication of the effect of infection on the host. -

WHO Immunological Basis for Immunization Series
WHO Immunological Basis for Immunization Series Module 21: Rotavirus Update 2019 Department of Immunization, Vaccines and Biologicals World Health Organization 20, Avenue Appia CH-1211 Geneva 27, Switzerland [email protected] http://www.who.int/immunization/en/ Immunization, Vaccines and Biologicals WHO Immunological Basis for Immunization Series Module 21: Rotavirus Update 2019 Immunization, Vaccines and Biologicals The immunological basis for immunization series: module 21: Rotavirus Vaccines (Immunological basis for immunization series; module 21) ISBN 978-92-4-000235-7 © World Health Organization 2020 Some rights reserved. This work is available under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons.org/licenses/by-nc-sa/3.0/igo). Under the terms of this licence, you may copy, redistribute and adapt the work for non-commercial purposes, provided the work is appropriately cited, as indicated below. In any use of this work, there should be no suggestion that WHO endorses any specific organization, products or services. The use of the WHO logo is not permitted. If you adapt the work, then you must license your work under the same or equivalent Creative Commons licence. If you create a translation of this work, you should add the following disclaimer along with the suggested citation: “This translation was not created by the World Health Organization (WHO). WHO is not responsible for the content or accuracy of this translation. The original English edition shall be the binding and authentic edition”. Any mediation relating to disputes arising under the licence shall be conducted in accordance with the mediation rules of the World Intellectual Property Organization. -

Wo 2007/056463 A2
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date PCT 18 May 2007 (18.05.2007) WO 2007/056463 A2 (51) International Patent Classification: (74) Agents: WILLIAMS, Kathleen et al.; Edwards Angell C12Q 1/70 (2006.01) C12Q 1/68 (2006.01) Palmer & Dodge LLP, P.O. Box 55874, Boston, MA 02205 (US). (21) International Application Number: (81) Designated States (unless otherwise indicated, for every PCT/US2006/043502 kind of national protection available): AE, AG, AL, AM, AT,AU, AZ, BA, BB, BG, BR, BW, BY, BZ, CA, CH, CN, (22) International Filing Date: CO, CR, CU, CZ, DE, DK, DM, DZ, EC, EE, EG, ES, FI, 9 November 2006 (09.1 1.2006) GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, KR, KZ, LA, LC, LK, LR, LS, (25) Filing Language: English LT, LU, LV,LY,MA, MD, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PG, PH, PL, PT, RO, RS, (26) Publication Language: English RU, SC, SD, SE, SG, SK, SL, SM, SV, SY, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW (30) Priority Data: (84) Designated States (unless otherwise indicated, for every 60/735,085 9 November 2005 (09. 11.2005) US kind of regional protection available): ARIPO (BW, GH, GM, KE, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, ZM, (71) Applicant (for all designated States except US): ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, TM), PRIMERA BIOSYSTEMS, INC. -

Pmtv, Pomovirus)
Gil et al. Actual Biol 33 (94): 69-84, 2011 CARACTERIZACIÓN GENOTÍPICA DE AISLAMIENTOS COLOMBIANOS DEL POTATO MOP-TOP VIRUS (PMTV, POMOVIRUS) GENOTYPIC CHARACTERIZATION OF COLOMBIAN ISOLATES OF POTATO MOP-TOP VIRUS (PMTV, POMOVIRUS) José F. Gil1, 4, Pablo A. Gutiérrez1, 5, José M. Cotes2, 6, Elena P. González3, 7, Mauricio Marín1, 8 Resumen Recientemente se detectó en Colombia la presencia del virus mop-top de la papa (PMTV, Pomovirus), transmitido por zoosporas del protozoo Spongospora subterranea f. sp. subterranea, agente causal de la sarna polvosa de la papa. Esta investigación se planteó con el fin de evaluar las características genotípicas de las cepas virales del PMTV obtenidas a partir de 20 muestras de suelos de cultivos de papa de los departamentos de Antioquia, Boyacá, Cundinamarca y Nariño. Para esto, se amplificaron mediante RT-PCR las regiones parciales que codifican para los genes de la cápside (CP) y del segundo gen del triple bloque de genes (TGB2), secuenciándose y comparándose con las secuencias de este virus depositadas en las bases de datos moleculares. Adicionalmente, se seleccionaron dos cepas del virus para aumentar el cubrimiento de las secuencias de los segmentos dos y tres (ARN 2 y ARN 3) del genoma viral. Los resultados reconfirmaron la presencia del virus PMTV en Colombia, detectándose dos variantes principales (I y II), una de ellas correspondiente al genotipo distribuido mundialmente (I); mientras que la otra variante (II) representa aislamientos virales de PMTV no registrados hasta ahora en otros países del mundo. Se espera que la información generada en esta investigación sea considerada en los programas de certificación de tubérculo semilla y del manejo cuarentenario de patógenos del cultivo de papa en el país. -

ICTV Code Assigned: 2011.001Ag Officers)
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the separate document “Help with completing a taxonomic proposal” Please try to keep related proposals within a single document; you can copy the modules to create more than one genus within a new family, for example. MODULE 1: TITLE, AUTHORS, etc (to be completed by ICTV Code assigned: 2011.001aG officers) Short title: Change existing virus species names to non-Latinized binomials (e.g. 6 new species in the genus Zetavirus) Modules attached 1 2 3 4 5 (modules 1 and 9 are required) 6 7 8 9 Author(s) with e-mail address(es) of the proposer: Van Regenmortel Marc, [email protected] Burke Donald, [email protected] Calisher Charles, [email protected] Dietzgen Ralf, [email protected] Fauquet Claude, [email protected] Ghabrial Said, [email protected] Jahrling Peter, [email protected] Johnson Karl, [email protected] Holbrook Michael, [email protected] Horzinek Marian, [email protected] Keil Guenther, [email protected] Kuhn Jens, [email protected] Mahy Brian, [email protected] Martelli Giovanni, [email protected] Pringle Craig, [email protected] Rybicki Ed, [email protected] Skern Tim, [email protected] Tesh Robert, [email protected] Wahl-Jensen Victoria, [email protected] Walker Peter, [email protected] Weaver Scott, [email protected] List the ICTV study group(s) that have seen this proposal: A list of study groups and contacts is provided at http://www.ictvonline.org/subcommittees.asp . -

Overlapping Genes Produce Proteins with Unusual Sequence Properties And
JVI Accepts, published online ahead of print on 29 July 2009 J. Virol. doi:10.1128/JVI.00595-09 Copyright © 2009, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights Reserved. 1 Overlapping genes produce proteins with unusual sequence properties and 2 offer insight into de novo protein creation 3 4 Corinne Rancurel1, Mahvash Khosravi2, Keith A. Dunker2, Pedro R. Romero2*, and David Karlin3* 5 6 1 Architecture et Fonction des Macromolécules Biologiques, Case 932, Campus de Luminy, 13288 Marseille 7 Cedex 9, France 8 2 Center for Computational Biology and Bioinformatics, 410 West 10th Street, Suite 5000, Indiana University 9 - Purdue University, Indianapolis, IN 46202-5122, USA, Downloaded from 10 3 25, rue de Casssis, 13008 Marseille, FRANCE 11 12 * Corresponding authors: [email protected], [email protected] 13 14 Running Title : Overlapping proteins have unusual sequence properties http://jvi.asm.org/ 15 Keywords 16 de novo gene creation; de novo protein creation; novel proteins; new proteins; orphan proteins; orphan 17 genes; ORFans; overlapping genes; overlapping reading frames; overprinting; unstructured proteins; 18 disordered proteins; intrinsic disorder; structural disorder; disorder prediction; profile-profile comparison; on April 22, 2020 by guest 19 PFAM; viral genomics; viral bioinformatics; viral structural genomics. 20 21 Abbreviations 22 Only abbreviations used in the main text are listed here; others are given in the figure captions. 23 30K, conserved domain of the 30K family of movement proteins; aa, amino acid; dsRNA, double-stranded 24 RNA; GT, guanylyltransferase; MP, movement protein; MT, methyltransferase; N, nucleoprotein; NSs, non 25 structural protein of orthobunyaviruses; ORF, open reading frame; PDB, protein databank (database of 26 protein structures); PFAM, Protein families (database of families of protein sequences); ssRNA, single- 27 stranded RNA; TGB, triple gene block; RE, relative (compositional) entropy; tm, transmembrane segment. -

Small Hydrophobic Viral Proteins Involved in Intercellular Movement of Diverse Plant Virus Genomes Sergey Y
AIMS Microbiology, 6(3): 305–329. DOI: 10.3934/microbiol.2020019 Received: 23 July 2020 Accepted: 13 September 2020 Published: 21 September 2020 http://www.aimspress.com/journal/microbiology Review Small hydrophobic viral proteins involved in intercellular movement of diverse plant virus genomes Sergey Y. Morozov1,2,* and Andrey G. Solovyev1,2,3 1 A. N. Belozersky Institute of Physico-Chemical Biology, Moscow State University, Moscow, Russia 2 Department of Virology, Biological Faculty, Moscow State University, Moscow, Russia 3 Institute of Molecular Medicine, Sechenov First Moscow State Medical University, Moscow, Russia * Correspondence: E-mail: [email protected]; Tel: +74959393198. Abstract: Most plant viruses code for movement proteins (MPs) targeting plasmodesmata to enable cell-to-cell and systemic spread in infected plants. Small membrane-embedded MPs have been first identified in two viral transport gene modules, triple gene block (TGB) coding for an RNA-binding helicase TGB1 and two small hydrophobic proteins TGB2 and TGB3 and double gene block (DGB) encoding two small polypeptides representing an RNA-binding protein and a membrane protein. These findings indicated that movement gene modules composed of two or more cistrons may encode the nucleic acid-binding protein and at least one membrane-bound movement protein. The same rule was revealed for small DNA-containing plant viruses, namely, viruses belonging to genus Mastrevirus (family Geminiviridae) and the family Nanoviridae. In multi-component transport modules the nucleic acid-binding MP can be viral capsid protein(s), as in RNA-containing viruses of the families Closteroviridae and Potyviridae. However, membrane proteins are always found among MPs of these multicomponent viral transport systems. -

Introduction to Viroids and Prions
Harriet Wilson, Lecture Notes Bio. Sci. 4 - Microbiology Sierra College Introduction to Viroids and Prions Viroids – Viroids are plant pathogens made up of short, circular, single-stranded RNA molecules (usually around 246-375 bases in length) that are not surrounded by a protein coat. They have internal base-pairs that cause the formation of folded, three-dimensional, rod-like shapes. Viroids apparently do not code for any polypeptides (proteins), but do cause a variety of disease symptoms in plants. The mechanism for viroid replication is not thoroughly understood, but is apparently dependent on plant enzymes. Some evidence suggests they are related to introns, and that they may also infect animals. Disease processes may involve RNA-interference or activities similar to those involving mi-RNA. Prions – Prions are proteinaceous infectious particles, associated with a number of disease conditions such as Scrapie in sheep, Bovine Spongiform Encephalopathy (BSE) or Mad Cow Disease in cattle, Chronic Wasting Disease (CWD) in wild ungulates such as muledeer and elk, and diseases in humans including Creutzfeld-Jacob disease (CJD), Gerstmann-Straussler-Scheinker syndrome (GSS), Alpers syndrome (in infants), Fatal Familial Insomnia (FFI) and Kuru. These diseases are characterized by loss of motor control, dementia, paralysis, wasting and eventually death. Prions can be transmitted through ingestion, tissue transplantation, and through the use of comtaminated surgical instruments, but can also be transmitted from one generation to the next genetically. This is because prion proteins are encoded by genes normally existing within the brain cells of various animals. Disease is caused by the conversion of normal cell proteins (glycoproteins) into prion proteins. -

Basic Properties of Viruses and Virus–Cell Interaction
WBV5 6/27/03 10:28 PM Page 49 Basic Properties of Viruses and Virus–Cell II Interaction PART ✷ VIRUS STRUCTURE AND CLASSIFICATION ✷ CLASSIFICATION SCHEMES ✷ THE BEGINNING AND END OF THE VIRUS REPLICATION CYCLE ✷ LATE EVENTS IN VIRAL INFECTION: CAPSID ASSEMBLY AND VIRION RELEASE ✷ HOST IMMUNE RESPONSE TO VIRAL INFECTION: THE NATURE OF THE VERTEBRATE IMMUNE RESPONSE ✷ PRESENTATION OF VIRAL ANTIGENS TO IMMUNE REACTIVE CELLS ✷ CONTROL AND DYSFUNCTION OF IMMUNITY ✷ MEASUREMENT OF THE IMMUNE REACTION ✷ STRATEGIES TO PROTECT AGAINST AND COMBAT VIRAL INFECTION ✷ VACCINATION — INDUCTION OF IMMUNITY TO PREVENT VIRUS INFECTION ✷ EUKARYOTIC CELL-BASED DEFENSES AGAINST VIRAL REPLICATION ✷ ANTIVIRAL DRUGS ✷ BACTERIAL ANTIVIRAL SYSTEMS — RESTRICTION ENDONUCLEASES ✷ PROBLEMS FOR PART II ✷ ADDITIONAL READING FOR PART II WBV5 6/27/03 10:28 PM Page 50 WBV5 6/27/03 3:35 PM Page 51 Virus Structure and Classification ✷ 5 Viral genomes ✷ Viral capsids ✷ Viral membrane envelopes ✷ CLASSIFICATION SCHEMES CHAPTER ✷ The Baltimore scheme of virus classification ✷ Disease-based classification schemes for viruses ✷ QUESTIONS FOR CHAPTER 5 Viruses are small compared to the wavelength of visible light; indeed, while the largest virus can be discerned in a good light microscope, viruses can only be visualized in detail using an electron microscope. A size scale with some important landmarks is shown in Fig. 5.1. Viruses are composed of a nucleic acid genomeor core, which is the genetic material of the virus, surrounded by a capsid made up of virus-encoded proteins. Viral genetic material encodes the structural proteins of the capsid and other viral proteins essential for other functions in initiating virus replication. The entire structure of the virus (the genome, the capsid, and — where present — the envelope) make up the virion or virus particle. -
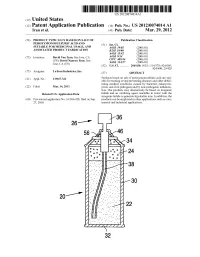
Us 2012/0074.014 A1 2 .. 1
US 2012O074O14A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0074.014 A1 Tran et al. (43) Pub. Date: Mar. 29, 2012 (54) PRODUCT TYPICALLY BASED ON SALT OF Publication Classification PEROXYMONOSULFURIC ACID AND (51) Int. Cl SUITABLE FOR MEDICINAL USAGE, AND iBio/02 (2006.01) ASSOCATED PRODUCT FABRICATION B23P 19/00 (2006.01) A633/42 (2006.01) (75) Inventors: David Van Tran, San Jose, CA A6IR 9/14 (2006.01) (US); David Nguyen Tran, San CD7C 409/244 (2006.01) Jose, CA (US) A6II 3/327 (2006.01) s (52) U.S. Cl. ............. 206/438: 562/1; 514/578; 424/605; 424/400; 29/428 (73) Assignee: LuTran Industries, Inc. (57) ABSTRACT 21) Appl. No.: 13AO47,742 Products based on salt of peroxyperoxVmonosulfuric acid are suit (21) Appl. No 9 able for treating or/and preventing diseases and other debili tating medical conditions caused by bacterial, eukaryotic, (22) Filed: Mar. 14, 2011 prion, and viral pathogens and by non-pathogenic inflamma tion. The products may alternatively be based on inorganic Related U.S. Application Data halide and an oxidizing agent reactable in water with the inorganic halide to generate hypohalite ions. In addition, the (60) Provisional application No. 61/386,928, filed on Sep. products can be employed in other applications such as com 27, 2010. mercial and industrial applications. 5T. 2 .. 2. 1.4. J., Patent Application Publication Mar. 29, 2012 Sheet 1 of 2 US 2012/0074014 A1 cro Q-D 22 Fig. 2a Fig.2b Patent Application Publication Mar. 29, 2012 Sheet 2 of 2 US 2012/0074014 A1 90 92 US 2012/0074014 A1 Mar. -

Evidence to Support Safe Return to Clinical Practice by Oral Health Professionals in Canada During the COVID-19 Pandemic: a Repo
Evidence to support safe return to clinical practice by oral health professionals in Canada during the COVID-19 pandemic: A report prepared for the Office of the Chief Dental Officer of Canada. November 2020 update This evidence synthesis was prepared for the Office of the Chief Dental Officer, based on a comprehensive review under contract by the following: Paul Allison, Faculty of Dentistry, McGill University Raphael Freitas de Souza, Faculty of Dentistry, McGill University Lilian Aboud, Faculty of Dentistry, McGill University Martin Morris, Library, McGill University November 30th, 2020 1 Contents Page Introduction 3 Project goal and specific objectives 3 Methods used to identify and include relevant literature 4 Report structure 5 Summary of update report 5 Report results a) Which patients are at greater risk of the consequences of COVID-19 and so 7 consideration should be given to delaying elective in-person oral health care? b) What are the signs and symptoms of COVID-19 that oral health professionals 9 should screen for prior to providing in-person health care? c) What evidence exists to support patient scheduling, waiting and other non- treatment management measures for in-person oral health care? 10 d) What evidence exists to support the use of various forms of personal protective equipment (PPE) while providing in-person oral health care? 13 e) What evidence exists to support the decontamination and re-use of PPE? 15 f) What evidence exists concerning the provision of aerosol-generating 16 procedures (AGP) as part of in-person -

A Tool for Hierarchical Clustering, Core Gene Detection and Annotation of (Prokaryotic) Viruses Cristina Moraru
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.14.448304; this version posted June 14, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. VirClust – a tool for hierarchical clustering, core gene detection and annotation of (prokaryotic) viruses Cristina Moraru Institute for Chemistry and Biology of the Marine Environment, Carl-von-Ossietzky –Str. 9 -11, D-26111 Oldenburg, Germany; [email protected] Abstract Recent years have seen major changes in the classification criteria and taxonomy of viruses. The current classification scheme, also called “megataxonomy of viruses”, recognizes five different viral realms, defined based on the presence of viral hallmark genes. Within the realms, viruses are classified into hierarchical taxons, ideally defined by their shared genes. Therefore, there is currently a need for virus classification tools based on such shared genes / proteins. Here, VirClust is presented – a novel tool capable of performing i) hierarchical clustering of viruses based on intergenomic distances calculated from their protein cluster content, ii) identification of core proteins and iii) annotation of viral proteins. VirClust groups proteins into clusters both based on BLASTP sequence similarity, which identifies more related proteins, and also based on hidden markow models (HMM), which identifies more distantly related proteins. Furthermore, VirClust provides an integrated visualization of the hierarchical clustering tree and of the distribution of the protein content, which allows the identification of the genomic features responsible for the respective clustering. By using different intergenomic distances, the hierarchical trees produced by VirClust can be split into viral genome clusters of different taxonomic ranks.