Biological Processes Modulating Longevity Across Primates
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mitochondrial Dysfunction in a Family with Psychosis and Chronic Fatigue Syndrome
ÔØ ÅÒÙ×Ö ÔØ Mitochondrial dysfunction in a family with psychosis and chronic fatigue syndrome Helena Torrell, Yolanda Alonso, Gl`oria Garrabou, David Mulet, Marc Catal´an, Alba Valiente-Pallej`a, Lidia Carre˜no-Gago, Elena Garc´ıa-Arum´ı, Elena Monta˜na, Elisabet Vilella, Lourdes Martorell PII: S1567-7249(16)30221-5 DOI: doi:10.1016/j.mito.2016.10.007 Reference: MITOCH 1139 To appear in: Mitochondrion Received date: 11 December 2015 Revised date: 7 July 2016 Accepted date: 26 October 2016 Please cite this article as: Torrell, Helena, Alonso, Yolanda, Garrabou, Gl`oria, Mulet, David, Catal´an, Marc, Valiente-Pallej`a, Alba, Carre˜no-Gago, Lidia, Garc´ıa-Arum´ı, Elena, Monta˜na, Elena, Vilella, Elisabet, Martorell, Lourdes, Mitochondrial dysfunc- tion in a family with psychosis and chronic fatigue syndrome, Mitochondrion (2016), doi:10.1016/j.mito.2016.10.007 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. ACCEPTED MANUSCRIPT Torrell et al, 1 de 42 Word count: 5190 Article title: Mitochondrial dysfunction in a family with psychosis and chronic fatigue syndrome Authors: Helena Torrell a§, Yolanda Alonso b, Glòria Garrabou c, David Mulet b, Marc Catalán c, Alba Valiente-Pallejà b, Lidia Carreño-Gago d, Elena García-Arumí d, Elena Montaña a, Elisabet Vilella a, Lourdes Martorell b§* Affiliations: a Centre for Omic Sciences, Centre Tecnòlogic de Nutrició i Salut. -

El Modernisme, Domènech I Montaner I El Seu Temps
. ISBN: 978-84-946247-4-2 112 HISTORIOGRAFIA DE REUS P. ANGUERA, J. MASSÓ, E. GORT, E. PEREA , M. FERRAN, J. NAVAIS Jornades d’arquitectura modernista 113 EL GUAITA – Articles periodístics CLAUDI ARNAVAT CARBALLIDO 114 PERE CAVALLÉ, CIUTADANIA REPUBLICANA XAVIER FERRÉ TRILL 115 UN DESIG POLÍTIC I UNS DELICTES POPULARS. L’AUDIÈNCIA CRIMINAL DE REUS El Modernisme, ALBERTO DIAZ FOZ 116 COMPLEMENTS CIRCUMSTANCIALS PERE ANGUERA NOLLA 117 BLOC DE TREBALL Domènech i Montaner SALVADOR JUANPERE 118 150 AUTÒGRAFS FONS CENTRE DE LECTURA LLIBRE DE SIGNATURES i el seu temps 119 LES CIUTATS DEL MÓN JOSEP IGLÉSIAS I ALTRES 120 HISTÒRIA DE REUS A TRAVÉS DEL BALL ALEIX CORT 121 AL COR DE LA MUNTANYA JOSEP CORNUDELLA 122 TRESCANT PER LA VIDA RAFEL FERRÉ MASIP 123 GOSAR PODER, SEMINARI GABRIEL FERRATER JORDI JULIÀ, XAVIERA MORÓS, RAMON GOMIS, JOSEP MURGADES, FINA MASDÉU 124 L’ENLLUMENAT PÚBLIC A LA CIUTAT DE REUS (1855-1965). DELS FANALS DE GAS A LES LÀMPADES ELÈCTRIQUES FLORENTÍ MOYANO I JIMÈNEZ 125 LA CRISI DE 1929 A REUS I COMARCA JOSEP BERTRAN CUDERS 126 INVASIÓ SUBTIL GUILLEM LATORRE 127 PREMSA I SOCIETAT 128 ERNEST CASAJUANA, POESIA I TEATRE ALBERT VENTURA 129 HISTÒRIES DEL SEGON PIS Taller d’escriptura del Centre de Lectura 130 UNA APOSTA NOVIOLENTA PER A LA PAU Trenta anys de Memorial Josep Vidal i Llecha 131 JORNADES D’ECONOMIA: CRISI ECONÒMICA I SOCIETAT. L’IMPACTE EN EL NOSTRE TERRITORI 132 LA LITERATURA COM A PATRIMONI I DESENVOLUPAMENT DEL TERRITORI TEMPS SEU I EL MODERNISME, DOMÈNECH I MONTANER EL 133 JORNADES LLIBRE ANTIC. LLIBRE ANTIC: MEMÒRIA DEL PASSAT Coordinadors 134 LA SARDANA BALLADA A REUS 135 UNS I ALTRES. -

1 L'hopital De La Santa Creu I De Sant Pau À Barcelona
L’HOPITAL DE LA SANTA CREU I DE SANT PAU À BARCELONA. PASSÉ, PRÉSENT ET FUTUR. Esther Balasch, Historienne de l’Art & Montserrat Caldés, Hôpital de la Santa Creu i de Sant Pau, Barcelona Lluís Domènech i Montaner Lluís Domènech i Montaner (1849-1923) est l’architecte des deux grands centres hospitaliers du modernisme (ou Art nouveau) catalan dont nous allons traiter au cours de cette conférence. Diplômé en sciences exactes, physiques et naturelles, il obtint en 1973 son titre d’architecte et se consacra à cette profession, ainsi qu’à l’enseignement à l’École d’Architecture, pendant 45 ans. Activiste politique, historien de l’art et homme de son temps, ses connaissances et études en minéralogie, zoologie et botanique sont déterminantes pour comprendre, en partie, l’ornementation de l’ensemble monumental de l’Hôpital de la Santa Creu i Sant Pau , qui constitue le complexe hospitalier le plus grand conçu en langage moderniste, avec l’Institut Pere Mata de Reus. Ce dernier, créé antérieurement par le même architecte, en 1897, sera le prédécesseur du futur HSCSP de Barcelone avec des réminiscences d’autres oeuvres de caractère civile, telles que le Palau de la Música Catalana de Barcelone. Institut Pere Mata de Reus L’Institut Pere Mata de Reus (Tarragone), dont le fondateur est le Dr. Emili Briansó Planas (1861-1922), occupe un terrain de 20 hectares et il est exclusivement réservé aux maladies mentales. À travers ce nouvel ensemble hospitalier, le Dr. Briansó Planas visait à réformer l’Hôpital psychiatrique de la province de Tarragone dans le but d’offrir une assistance adéquate aux personnes souffrant de maladies mentales diverses. -

Medical List
U.S. Consulate General Paseo Reina Elisenda 23 – 08034 Barcelona, Spain Tel: (34) 93-280-2227 – Fax: (34) 93-280-6175 Email: [email protected] Web Site: https://es.usembassy.gov/embassy-consulates/barcelona/ _________________________________________________________________________________________________________________________________ Medical List This list is prepared by the Consular Section of the Consulate General of the United States of America in Barcelona, Spain. This list covers the U.S. Consulate General’s consular district consisting of: The Autonomous Community of Catalonia; Provinces of Barcelona, Lleida, Tarragona and Girona, The Autonomous Community of Aragon; Provinces of Zaragoza, Huesca and Teruel, and the Principality of Andorra. DISCLAIMER: The U.S. Consulate General in Barcelona, Spain assumes no responsibility or liability for the professional ability or reputation of, or the quality of services provided by, the medical professionals, medical facilities or air ambulance services whose names appear on the following list. Inclusion on this list is in no way an endorsement by the Department of State or the U.S. Consulate General. Names are listed alphabetically, and the order in which they appear has no other significance. The information in the list on professional credentials and areas of expertise is provided directly by the medical professional, medical facility or air ambulance service; the Consulate General is not in a position to vouch for such information. You may receive additional information about the individuals and facilities on the list by contacting local medical boards and associations (or its equivalent) or local licensing authorities. Good medical care is available in Spain. However, regulations regarding medications vary from those in the United States. -

A Non-Interventional Naturalistic Study of the Prescription Patterns of Antipsychotics in Patients with Schizophrenia from the Spanish Province of Tarragona
RESEARCH ARTICLE A Non-Interventional Naturalistic Study of the Prescription Patterns of Antipsychotics in Patients with Schizophrenia from the Spanish Province of Tarragona Ana M. Gaviria1☯, José G. Franco1☯*, Víctor Aguado2, Guillem Rico2, Javier Labad1, Joan de Pablo1, Elisabet Vilella1 1 Hospital Universitari Institut Pere Mata, Universitat Rovira i Virgili, CIBERSAM, IISPV Reus, Spain, 2 Hospital Universitari Institut Pere Mata, Reus, Spain ☯ These authors contributed equally to this work. * [email protected] Abstract OPEN ACCESS Citation: Gaviria AM, Franco JG, Aguado V, Rico G, Labad J, de Pablo J, et al. (2015) A Non- Background Interventional Naturalistic Study of the Prescription The analysis of prescribing patterns in entire catchment areas contributes to global mapping Patterns of Antipsychotics in Patients with Schizophrenia from the Spanish Province of of the use of antipsychotics and may improve treatment outcomes. Tarragona. PLoS ONE 10(10): e0139403. doi:10.1371/journal.pone.0139403 Objective Editor: Peter John McKenna, Benito Menni Complejo To determine the pattern of long-term antipsychotic prescription in outpatients with schizo- Asistencial en Salud Mental, SPAIN phrenia in the province of Tarragona (Catalonia-Spain). Received: April 27, 2015 Accepted: September 10, 2015 Methods Published: October 1, 2015 A naturalistic, observational, retrospective, non-interventional study based on the analysis Copyright: © 2015 Gaviria et al. This is an open of registries of computerized medical records from an anonymized database of 1,765 access article distributed under the terms of the patients with schizophrenia treated between 2011 and 2013. Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any Results medium, provided the original author and source are credited. -

Die Top-5 Sehenswürdigkeiten Von Reus
1 Katalonien Tourismus Reus Eva Hakes · Friday, June 28th, 2019 Überblick Es gibt viele Anlässe, für einen Ausflug nach Reus. Der Klassiker unter den Beweggründen für einen Besuch der Geburtsstadt Antoni Gaudís ist Regenwetter in nahegelegenen Strandurlaubs-Orten wie Cambrils oder Salou. Selbstverständlich ist schlechtes Strandwetter ein legitimer Grund sich der Kultur zu widmen. Allerdings lohnt ein Besuch der Stadt Reus sich definitiv auch bei strahlendem Sonnenschein. Das Institut Pere Mata ist eines der Highlights von Reus © Ajuntament de Reus Reus, die Stadt des Modernisme Reus hütet etwa 80 beeindruckende modernistische Bauwerke. 29 von ihnen sind Teil der Modernisme-Route von Reus. Interessanterweise stammt keines dieser Bauwerke vom in Reus geborenen Gaudí, der in so unnachahmlicher Weise Barcelona geprägt Katalonien Tourismus - 1 / 10 - 02.07.2021 2 hat. Dafür finden sich hier u.a. Werke von Domènech i Montaner, die zu den Höhepunkten der modernistichen Architektur Kataloniens zählen. Zu diesen gehören zum Beispiel die Casa Navàs und das Institut Pere Mata, ein Vorläufer des berühmten Hospital de Sant Pau in Barcelona, das zum Unesco-Welterbe zählt. Neben Bauwerken von Domènech i Montaner kann man auf der Modernisme-Route auch den katalanischen Jugendstil in seiner Ausprägung durch Architekten wie Pere Caselles und Joan Rubió i Bellver kennenlernen. Detail der Casa Navàs an der Plaça del Mercadal © Miguel Raurich Während Gaudí zu Beginn des 20. Jahrhunderts in Reus offenbar weit weniger Anerkennung erhielt als seine Kollegen, hat ihm seine Heimatstadt zu Beginn des 21. Jahrhunderts ein grandioses Denkmal gesetzt. Das 2007 eröffnete Gaudí Centre gewährt mit einem multimedialen und multisensorialen Ansatz faszinierende Einblicke in das Leben und Schaffen des Ausnahme-Architekten. -

Mosaic and Trencadís in the Palau De La Música Catalana Cristina Martí Robledo A*
Available online at www.IJournalSE.org Emerging Science Journal Vol. 2, No. 2, April, 2018 Mosaic and Trencadís in the Palau De La Música Catalana Cristina Martí Robledo a* a Restauracions POLICROMIA SL, Catalonia Abstract Keywords: Modernism has provided Catalonia with buildings of great artistic value, such as the Palau de la Música Modernisme; Catalana located in the heart of Barcelona. Its architect, Lluís Domènech i Montaner, approached Lluís Domènech i Montaner; ornamentation as a thorough integration of arts and crafts: ceramic is treated as the ¨key¨ element, Conservation; decorating the entire building while coating and protecting its internal metallic structure at the same Restauration. time. During Modernisme, the classic mosaic technique, based on the use of small tiles (or tesserae) made Article History: of ceramics or glass paste, incorporated a new method called trencadís (A procedure carried out using irregular pieces of tiles fragmented randomly and put together in an abstract way). Received: 20 March 2018 Accepted: 01 May 2018 1- Introduction In the heart of Barcelona's old town (Ciutat Vella) lies the emblematic Palau de la Música, a building from the Modernista era, which was declared a BCIN (Heritage Site of National Interest) in 1971 and UNESCO World Heritage Site in 1997. (Figure 1). It is located on the junction of the streets Sant Pere més Alt, Amadeus Vives and Sant Francesc de Paula, where there once stood a Convent with the same name, and which in 1992 was knocked down. Its architect, Lluís Domènech i Montaner was one of a series of great names that the Modernista era produced. -

Domènech I Montaner a Reus
DOMÈNECH I MONTANER A REUS DUES OBRES SINGULARS: INSTITUT PERE MATA I CASA NAVÀS Paula Farrés Arandía NIUB: 16465083 Tutora: Dra. Teresa M. Sala Garcia Grau d’Història de l’Art Bloc de Segle XIX i Primeres Avantguardes Universitat de Barcelona Curs: 2017-2018 Sempre que una idea organitzadora domina un poble, sempre que esclata una nova civilització, apareix una nova època artística. Lluís Domènech i Montaner, “En busca d’una arquitectura nacional”, La Renaixença (1878). 1 Índex 1. Introducció ................................................................................................................................................... 3 1.1. Objectius .............................................................................................................................................. 4 1.2. Estat de la qüestió ................................................................................................................................ 5 1.3. Metodologia ....................................................................................................................................... 10 2. Context de la fi de segle: el fonament de l’Art Noveau Internacional ....................................................... 12 2.1. El projecte cultural del Modernisme ................................................................................................. 13 2.2. Arquitectura del Modernisme i la ciutat de Barcelona ...................................................................... 16 2.3. El Modernisme a Reus ...................................................................................................................... -

Language Policy Report 2010
LANGUAGE POLICY REPORT 2010 I. The legal framework for language I. THE LEGAL FRAMEWORK FOR LANGUAGE 1. Prevailing legal framework 1.1. The Statute of Autonomy of Catalonia 1.1.1. The constitutional and statutory framework of the system of official languages and the powers of the Generalitat in this area 1.1.2. Language rights and duties and the guiding principles of public policy with respect to language 1.1.2.1. Public Administration and associated bodies 1.1.2.2. Education 1.1.2.3. The media 1.1.2.4. The socioeconomic sphere 1.1.2.5. Cooperation and external promotion 1.1.3. The recognition of Catalan sign language 1.2. The Language Policy Act 1.2.1. Public Administration and associated bodies 1.2.1.1. Regulation of use 1.2.1.2. The regulation of knowledge 1.2.1.3. The use of Catalan in the field of justice 1.2.1.4. Registries and notaries 1.2.1.5. Onomastics 1.2.2. Education 1.2.2.1. Compulsory education 1.2.2.2. University education 1.2.3. Media and cultural industries 1.2.3.1. The broadcast media 1.2.3.2. The Catalan Broadcasting Corporation 1.2.3.3. Local TV stations 1.2.3.4. The Catalan Audiovisual Council 1.2.3.5. Information and communication technologies 1.2.3.6. The cultural sphere 1.2.4. The socioeconomic sphere 1.2.5. External promotion 1 I. The legal framework for language 1.3. Education Act 1.4. Consumer Code of Catalonia Act 1.4.1. -

Màster Erasmus Mundus En Vi I Turisme, Organitzat Amb URV-SOCIETAT Les Universitats De Porto I Bordeus I Coordinat Per 32 La Universitat La Nostra Universitat
// Màster Erasmus Mundus REVISTA DE LA UNIVERSITAT ROVIRA I VIRGILI ROVIRA DE LA UNIVERSITAT REVISTA , en Vi i Turisme URV // 11 / 15 n24 JOAN PEDREROL: DOCÈNCIA: URV-SOCIETAT: “EL TERRITORI SE APOSTA NEIX LA CÀTEDRA SENT ORGULLÓS PEL MÀSTER DE DOLOR INFANTIL DE LA UNIVERSITAT” REVISTA DE LA UNIVERSITAT ROVIRA I VIRGILI 11 / 15 · N24 EDITA Universitat Rovira i Virgili DIRECCIÓ Ignasi Soler REDACCIÓ Berta Ramos, Montse Cartañà, Cristina Mallo, Cinta S. Bellmunt, Xavier Jiménez, Arnau Marín, Oriol Margalef, Roser Royo, Rosa Garcia, Jaume Llambrich i Javi Polinario Gabinet de Comunicació i Relacions Externes C/ de l’Escorxador, s/n 43003 Tarragona Tel. 977 558006 Fax 977 558258 [email protected] ASSESSORAMENT LINGÜÍSTIC Servei Lingüístic de la URV FOTOGRAFIA Ramon Torrents Xavi Jurio Francesc Nadal Vicent Pellicer Rafael López-Monné MAQUETACIÓ Feeling Comunicació GESTIÓ I DISTRIBUCIÓ Jordi Garcia DIPÒSIT LEGAL: B – 48078 – 2004 facebook.com/universitatroviraivirgili twitter.com/universitatURV urv és una publicació de distribució gratuïta // Sumari PERFILS El valor del 4 Sara Gomis: “Cal professionalitzar el sector turístic” màster El màster s’ha convertit en una prioritat de les SUMARI institucions d’educació superior fonamentades en INSTITUCIÓ la generació de coneixement, tant per la formació de 8 La URV al món qualitat que s’aporta a la societat com per la connexió amb el doctorat i la recerca. Aquest és també el camí que hem seguit des de la URV, amb una forta aposta pels màsters en aquesta doble funció formativa i de recerca, i amb la derivada de captació de talent que comporten. RECERCA En aquest curs acadèmic 2015-16 que acabem d’iniciar 13 Protegint la ens congratulem que els estudiants de màster de la privacitat a nostra universitat hagin augmentat més d’un 13%, un les xarxes increment important que significa assolir la xifra de 1.200 persones matriculades. -

Satisfaction with Life Domains Scale by Baker and Intagliata
Available online at www.sciencedirect.com Comprehensive Psychiatry 50 (2009) 76–80 www.elsevier.com/locate/comppsych Adaptation and validation of the quality-of-life scale: Satisfaction with Life Domains Scale by Baker and Intagliata ⁎ Janina Carlsona,1, Susana Ochoaa, ,1, Josep Maria Haroa,1, Gemma Escartína,b,1, Maribel Ahuirc,1, Alfonso Gutierrez-Zotesd,1, Manel Salameroe,1, Joaquim Valerod,1, Silvia Cañizaresf,1, Miquel Bernardog,1, Jose Cañetef,1, Pedro Gallob,1 aResearch and Developmental Unit of Sant Joan de Déu-SSM, Fundació Sant Joan de Déu, 08330 Sant Boi de Llobregat, Barcelona, Spain bFundació Seny, Barcelona, Spain cFundació Clínic, Hospital Clínic de Barcelona, IDIBAPS, Barcelona, Spain dHospital Psiquiàtric Universitari Institut Pere Mata i Unitat de Psiquiatria, Universitat Rovira Virgili, Reus, Spain eInstitut Clínic de Neurociencias, Servicio de Psiquiatría y Psicología Clínica, Hospital Clínic, Universitat de Barcelona, Barcelona, Spain fHospital de Mataró, Mataró, Spain gHospital Clínic Barcelona, Universidad de Barcelona, IDIBAPS, Barcelona, Spain Abstract Objectives: There are few quality-of-life instruments specifically for schizophrenia; thus, the objective of our study is to adapt and validate the Satisfaction with Life Domains Scale (SLDS) by Baker and Intagliata. Method: This is a validation study in which the subjects were evaluated on 2 occasions (24-48 hours). The sample is composed of people with schizophrenia from 18 to 65 years old and who were seen in one of the following centers: Sant Joan de Déu-SSM, Hospital Clínic, Hospital de Mataró and Hospital Pere Mata. The SLDS was administered, along with Positive and Negative Syndrome Scale, the Clinical General Impression for Schizophrenia, Global Assessment of Function, Disability Assessment Scale—short version, Beck Cognitive Insight, and the Strauss and Carpenter Prognostic Scale. -
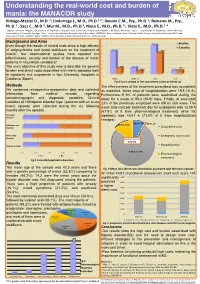
Understanding the Real-World Cost and Burden of Mania: the MANACOR Study Hidalgo-Mazzei D., M.D
Understanding the real-world cost and burden of mania: the MANACOR study Hidalgo-Mazzei D., M.D. 1; Undurraga J., M.D., Ph.D.1 2 ; Bonnin C.M., Psy., Ph.D. 1; Reinares M., Psy., Ph.D. 1 , Saez C., M.D 3, Mur M., M.D., Ph.D.4, Nieto E., M.D., Ph.D. 5; Vieta E., M.D., Ph.D.1 * 1 Bipolar Disorders Program, Department of Psychiatry, Hospital Clinic, University of Barcelona, IDIBAPS, CIBERSAM, Barcelona, Spain. 2 Department of Psychiatry, Clínica Alemana, Universidad del Desarrollo, Santiago, Chile. 3 University Psychiatric Hospital, Institut Pere Mata, CIBERSAM, Reus, Catalonia, Spain 4 Mental Health Service, Santa Maria Hospital, IRB LLeida, University of LLeida, Catalonia, Spain. 5 Althaia, Xarxa Assistencial Universitária de Manresa, Catalonia, Spain. Background and Aims Baseline Even though the results of clinical trials show a high efficacy 40 6 months of antipsychotics and mood stabilizers on the treatment of 35 -26% mania1, few observational studies have reported the 30 effectiveness, security and burden of the disease of manic 25 patients in naturalistic conditions2. 20 15 The main objective of this study was to describe the general -75% 10 -3% burden and direct costs associated with manic episodes both -52% 5 in inpatients and outpatients in four University Hospitals in 0 Catalonia (Spain). YMRS HDRS-17 FAST SCIP Fig.3 Score change of the assessment scales at follow-up Methods The effectiveness of the treatment prescribed was acceptable We combined retrospective-prospective data and collected as expected. Mean days of hospitalization were 19.8 (11.6). information from medical records regarding Furthermore,11.9% of patients were readmitted during the sociodemographic and diagnostic variables.