Facial Genetics: a Brief Overview
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Clinical Review Nursingingeneralpractice
The health benefits of nose breathing Item Type Article Authors Allen, Ruth Publisher Nursing in General Practice Journal Nursing in General Practice Download date 01/10/2021 07:15:20 Link to Item http://hdl.handle.net/10147/559021 Find this and similar works at - http://www.lenus.ie/hse clinical review nursingingeneralpractice The health benefits of nose breathing DR Alan RUth, BehaviouRal Medicine PRactitioneR “For breath is life, and if you breathe well you will live long on earth.” sanskrit Proverb For the most part people are unaware of their breathing and take it for granted that they do it correctly. t has been estimated that approximately one third of people ing. However, it has been estimated that up to 30-50% of modern don’t breathe well enough to sustain normal health. These adults breathe through the mouth, especially during the early people do not get enough oxygenation of their cells, tissues morning hours. and organs. In the book Behavioural and Psychological Ap- Mouth breathing is common in individuals whose nasal proaches to Breathing Disorders, Dr Chandra Patel describes passages are blocked or restricted. A deviated nasal septum Ithe problem with breathing as follows: or small nostril size can lead a person to breathe through their “We start life with a breath, and the process continues mouth instead of their nose. However, breathing through the automatically for the rest of our lives. Because breathing mouth most of the time was not nature’s intention. Many studies continues on its own, without our awareness, it does not have demonstrated that chronic mouth breathing can result in a necessarily mean that it is always functioning for optimum number of adverse health consequences (see Table 1). -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Chromosome 14 Transfer and Functional Studies Identify a Candidate Tumor Suppressor Gene, Mirror Image Polydactyly 1, in Nasopharyngeal Carcinoma
Chromosome 14 transfer and functional studies identify a candidate tumor suppressor gene, Mirror image polydactyly 1, in nasopharyngeal carcinoma Arthur Kwok Leung Cheunga, Hong Lok Lungb, Josephine Mun Yee Kob, Yue Chenga,c, Eric J. Stanbridged, Eugene R. Zabarovskye, John M. Nichollsf, Daniel Chuab, Sai Wah Tsaog, Xin-Yuan Guanb, and Maria Li Lungb,1 aDepartment of Biology and Center for Cancer Research, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong (SAR), People’s Republic of China; bDepartment of Clinical Oncology, University of Hong Kong, Pokfulam, Hong Kong (SAR), People’s Republic of China; cDepartment of Biology, City of Hope, Beckman Research Institute, Duarte,CA 91010; dDepartment of Microbiology and Molecular Genetics, University of California, Irvine, CA 92697; eDepartment of Microbiology, Tumor and Cell Biology, Karolinska Institute, Stockholm, Sweden; and Departments of fPathology and gAnatomy, University of Hong Kong, Pokfulam, Hong Kong (SAR), People’s Republic of China Edited by George Klein, Karolinska Institutet, Stockholm, Sweden, and approved July 7, 2009 (received for review January 7, 2009) Chromosome 14 allelic loss is common in nasopharyngeal carcinoma particular interest to us that chromosome 14 loss is associated with (NPC) and may reflect essential tumor suppressor gene loss in tumor- cancer metastasis in breast tumors (15) and with poor clinical prognosis igenesis. An intact chromosome 14 was transferred to an NPC cell line for other head and neck cancers (16). Thus, it is possible that a using a microcell-mediated chromosome transfer approach. Microcell chromosome 14 TSG may be a useful prognostic marker in NPC. hybrids (MCHs) containing intact exogenously transferred chromo- In this study, we obtained functional evidence showing definitively some 14 were tumor suppressive in athymic mice, demonstrating that that chromosome 14 is tumor suppressive in NPC. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Empty Nose Syndrome
EMPTY NOSE SYNDROME 1 Empty Nose Syndrome 1.1 Definition The descriptive term “Empty nose syndrome” (ENS) was first coined by Kern and Stenkvist in 1994, who described the condition with substantial loss of tissue inside the nose in the region of the inferior and middle turbinates as “empty nose” (Scheithauer, 2010). “Empty nose” patients suffer from chronic nasal crusting, dryness, and intermittent bleeding. The most distinctive symptom of ENS is “paradoxical obstruction”- in that the patient suffers from difficulty breathing despite a widely patent nasal airway (Sozansky & Houser, 2014). ENS patients describe their feeling of nasal obstruction as constant and continuous: feeling of suffocation, inability or significant difficulty to breathe through their nose, feeling that their nose is too open, sensation of excessive airflow, shortness of breath, difficulty to properly inflate the lungs, lack of nasal resistance, and/or undifferentiated breathing difficulties (Houser, 2006; Houser, 2007; Chhabra & Houser, 2009; Scheithauer, 2010; Coste et al, 2012). 1.2 Primary symptoms ENS patients may experience various debilitating and devastating symptoms, in varying degrees, that can occur after excessive resection of functioning turbinate tissue from the nasal cavity (Moore & Kern, 2001). While the primary symptom is nasal airway obstruction, other symptoms depend upon the type of surgery, extent of functional nasal tissue removal, nature of the bacterial organisms growing inside the nasal cavity, and other unidentified factors. The set and level of severity of symptoms depends upon each individual case. ENS-related symptoms can be divided into three major categories and may include: Respiratory: paradoxical obstruction (feeling of suffocation, inability or significant difficulty to breathe through the nose, feeling that the nose is too open, sensation of excessive airflow, lack of nasal resistance, shortness of breath (dyspnea), difficulty to properly inflate the lungs, difficulty drawing a full breath, weakened airflow, and undifferentiated breathing difficulties) 1. -

The Facebase Consortium: a Comprehensive Program To
Manuscript Click here to view linked References The FaceBase Consortium: A Comprehensive Program to Facilitate Craniofacial Research Harry Hochheiser1*, Bruce J. Aronow2, Kristin Artinger3, Terri H.Beaty4, James F. Brinkley5, Yang Chai6, David Clouthier3, Michael L. Cunningham7, Michael Dixon8, Leah Rae Donahue9, Scott E. Fraser10, Junichi Iwata6, Mary L. Marazita11, Jeffrey C. Murray12, Stephen Murray9, John Postlethwait13, Steven Potter14, Linda Shapiro5, Richard Spritz15, Axel Visel16, Seth M. Weinberg17 and Paul A. Trainor18*, for the FaceBase Consortium. 1. Department of Biomedical Informatics, University of Pittsburgh, Pittsburgh PA 15232 USA 2. Divisions of Biomedical Informatics and Developmental Biology, Center for Computational Medicine, Cincinnati Children's Hospital Medical Center, University of Cincinnati College of Medicine, CHRF 8504, 3333 Burnet Ave Cincinnati, OH 45229 USA 3. Department of Craniofacial Biology, University of Colorado Denver Anschutz Medical Campus, Aurora, CO 80045 4. Department of Epidemiology, Johns Hopkins University, 615 N. Wolfe Street Baltimore, MD. 21205 USA 5. Department of Computer Science and Engineering, University of Washington, Box 352350 Seattle, WA 98195-2350 USA 6. Center for Craniofacial Molecular Biology, Ostrow School of Dentistry, University of Southern California, 2250 Alcazar Street, CSA 103, Los Angeles, CA 90033 7. Seattle Children’s Hospital, 4800 Sand Point Way NE, Seattle, WA 98105 8. Faculty of Medical and Human Sciences, Manchester Academic Health Sciences Centre, and Faculty of Life Sciences, Michael Smith Building, University of Manchester, Oxford Road, Manchester, M13 9PT, England 1 9. Jackson Laboratory, 600 Main St., Bar Harbor, ME 04609 USA 10. Biological Imaging Center Beckman Institute 133, M/C 139-74 California Institute of Technology Pasadena, CA 91125 11. -
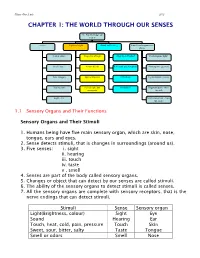
Chapter 1: the World Through Our Senses
Science Form 2 note 2012 CHAPTER 1: THE WORLD THROUGH OUR SENSES The World through our senses senses Light and sight Sound and hearing Stimuli and responses in plants Touch (skin) Properties of light Properties of sound Phototropism (light) Smell (nose) Vision defects Reflection and absorption Geotropism (gravity) Taste (tongue) Optical illusions limitations Hydrotropism (water) Hearing (ear) Stereoscopic and stereophonic Thigmotropism (move monocular toward) Sight (eye) Nastic movement (move run away) 1.1 Sensory Organs and Their Functions Sensory Organs and Their Stimuli 1. Humans being have five main sensory organ, which are skin, nose, tongue, ears and eyes. 2. Sense detects stimuli, that is changes in surroundings (around us). 3. Five senses: i. sight ii. hearing iii. touch iv. taste v . smell 4. Senses are part of the body called sensory organs. 5. Changes or object that can detect by our senses are called stimuli. 6. The ability of the sensory organs to detect stimuli is called senses. 7. All the sensory organs are complete with sensory receptors, that is the nerve endings that can detect stimuli. Stimuli Sense Sensory organ Light(Brightness, colour) Sight Eye Sound Hearing Ear Touch, heat, cold, pain, pressure Touch Skin Sweet, sour, bitter, salty Taste Tongue Smell or odors Smell Nose Science Form 2 note 2012 Laman web. http://freda.auyeung.net/5senses/see.htm http://freda.auyeung.net/5senses/touch.htm http://freda.auyeung.net/5senses/hear.htm http://freda.auyeung.net/5senses/taste.htm http://freda.auyeung.net/5senses/smell.htm 1.2 The Pathway from Stimulus to Response PMR 05 Stimulus Sensory organs Nerves Brain Nerve Response Figure 1.2 The summary of the pathway from stimulus to response 1. -

A Dissertation Entitled the Androgen Receptor
A Dissertation entitled The Androgen Receptor as a Transcriptional Co-activator: Implications in the Growth and Progression of Prostate Cancer By Mesfin Gonit Submitted to the Graduate Faculty as partial fulfillment of the requirements for the PhD Degree in Biomedical science Dr. Manohar Ratnam, Committee Chair Dr. Lirim Shemshedini, Committee Member Dr. Robert Trumbly, Committee Member Dr. Edwin Sanchez, Committee Member Dr. Beata Lecka -Czernik, Committee Member Dr. Patricia R. Komuniecki, Dean College of Graduate Studies The University of Toledo August 2011 Copyright 2011, Mesfin Gonit This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. An Abstract of The Androgen Receptor as a Transcriptional Co-activator: Implications in the Growth and Progression of Prostate Cancer By Mesfin Gonit As partial fulfillment of the requirements for the PhD Degree in Biomedical science The University of Toledo August 2011 Prostate cancer depends on the androgen receptor (AR) for growth and survival even in the absence of androgen. In the classical models of gene activation by AR, ligand activated AR signals through binding to the androgen response elements (AREs) in the target gene promoter/enhancer. In the present study the role of AREs in the androgen- independent transcriptional signaling was investigated using LP50 cells, derived from parental LNCaP cells through extended passage in vitro. LP50 cells reflected the signature gene overexpression profile of advanced clinical prostate tumors. The growth of LP50 cells was profoundly dependent on nuclear localized AR but was independent of androgen. Nevertheless, in these cells AR was unable to bind to AREs in the absence of androgen. -

Toxicological Profile for Acetone Draft for Public Comment
ACETONE 1 Toxicological Profile for Acetone Draft for Public Comment July 2021 ***DRAFT FOR PUBLIC COMMENT*** ACETONE ii DISCLAIMER Use of trade names is for identification only and does not imply endorsement by the Agency for Toxic Substances and Disease Registry, the Public Health Service, or the U.S. Department of Health and Human Services. This information is distributed solely for the purpose of pre dissemination public comment under applicable information quality guidelines. It has not been formally disseminated by the Agency for Toxic Substances and Disease Registry. It does not represent and should not be construed to represent any agency determination or policy. ***DRAFT FOR PUBLIC COMMENT*** ACETONE iii FOREWORD This toxicological profile is prepared in accordance with guidelines developed by the Agency for Toxic Substances and Disease Registry (ATSDR) and the Environmental Protection Agency (EPA). The original guidelines were published in the Federal Register on April 17, 1987. Each profile will be revised and republished as necessary. The ATSDR toxicological profile succinctly characterizes the toxicologic and adverse health effects information for these toxic substances described therein. Each peer-reviewed profile identifies and reviews the key literature that describes a substance's toxicologic properties. Other pertinent literature is also presented, but is described in less detail than the key studies. The profile is not intended to be an exhaustive document; however, more comprehensive sources of specialty information are referenced. The focus of the profiles is on health and toxicologic information; therefore, each toxicological profile begins with a relevance to public health discussion which would allow a public health professional to make a real-time determination of whether the presence of a particular substance in the environment poses a potential threat to human health. -

A High-Resolution 3D Epigenomic Map Reveals Insights Into the Creation of the Prostate Cancer Transcriptome
ARTICLE https://doi.org/10.1038/s41467-019-12079-8 OPEN A high-resolution 3D epigenomic map reveals insights into the creation of the prostate cancer transcriptome Suhn Kyong Rhie 1, Andrew A. Perez1, Fides D. Lay1, Shannon Schreiner 1, Jiani Shi1, Jenevieve Polin 1 & Peggy J. Farnham 1 1234567890():,; To better understand the impact of chromatin structure on regulation of the prostate cancer transcriptome, we develop high-resolution chromatin interaction maps in normal and pros- tate cancer cells using in situ Hi-C. By combining the in situ Hi-C data with active and repressive histone marks, CTCF binding sites, nucleosome-depleted regions, and tran- scriptome profiling, we identify topologically associating domains (TADs) that change in size and epigenetic states between normal and prostate cancer cells. Moreover, we identify normal and prostate cancer-specific enhancer-promoter loops and involved transcription factors. For example, we show that FOXA1 is enriched in prostate cancer-specific enhancer- promoter loop anchors. We also find that the chromatin structure surrounding the androgen receptor (AR) locus is altered in the prostate cancer cells with many cancer-specific enhancer-promoter loops. This creation of 3D epigenomic maps enables a better under- standing of prostate cancer biology and mechanisms of gene regulation. 1 Department of Biochemistry and Molecular Medicine and the Norris Comprehensive Cancer Center, Keck School of Medicine, University of Southern California, Los Angeles, CA 90089, USA. Correspondence and requests for materials should be addressed to S.K.R. (email: [email protected]) or to P.J.F. (email: [email protected]) NATURE COMMUNICATIONS | (2019) 10:4154 | https://doi.org/10.1038/s41467-019-12079-8 | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-019-12079-8 rostate cancer is the leading cause of new cancer cases and Results Pthe second most common cancer in men and the fourth Cancer-specific TADs that lead to transcriptome changes. -
Investigating the Practical Use of Computational Fluid Dynamics Simulation of Airflow in the Nasal Cavity and Paranasal Sinuses NOSE Version 1.0 2018-08-30
NOSE Pilot Study Investigating the Practical Use of Computational Fluid Dynamics Simulation of Airflow in the NNOSEasal Cavity and Paranasal Sinuses NOSE Pilot Study Investigating the Practical Use of Computational Fluid Dynamics Simulation of Airflow in the Nasal Cavity and Paranasal Sinuses NOSE Version 1.0 2018-08-30 NOSE Pilot Study Investigating the Practical Use of Computational Fluid Dynamics Simulation Version 1.0 of Airflow in the Nasal Cavity and Paranasal Sinuses NOSE Pilot Study Investigating the Practical Use of Computational Fluid Dynamics Simulation of Airflow in the Nasal Cavity and Paranasal Sinuses Project Number 1000043433 Project Title NOSE Pilot Study Document Reference Investigating the Practical Use of Computational Date 2018-08-30 Title Fluid Dynamics Simulation of Airflow in the Nasal Cavity and Paranasal Sinuses Document Name NOSE_PS_FINAL_v1 Version Draft draft final Restrictions public internal restricted : Distribution Steirische Forschungsförderung Authors Koch Walter, Koch Gerda, Vitiello Massimo, Ortiz Ramiro, Stockklauser Jutta, Benda Odo Abstract The NOSE Pilot study evaluated the technical and scientific environment required for establishing a service portfolio that includes CFD simulation and 3D visualization services for ENT specialists. For this purpose the state-of-the-art of these technologies and their use for upper airways diagnostics were analysed. Keywords Rhinology, Computational Fluid Dynamics, 3D Visualization, Clinical Pathways, Service Center, Knowledge Base Document Revisions Version Date Author(s) Description of Change 1.0 2018-08-30 Koch Walter, Koch Final Version Gerda, Vitiello Massimo, Ortiz NOSERamiro, Stockklauser Jutta, Benda Odo 2018-08-30 Seite 3 / 82 Copyright © AIT ForschungsgesmbH NOSE_PS_FINAL_v1 NOSE Pilot Study Investigating the Practical Use of Computational Fluid Dynamics Simulation Version 1.0 of Airflow in the Nasal Cavity and Paranasal Sinuses Table of Contents Acknowledgments ..................................................................................