Branched-Chain Ester Biosynthesis in Ripening Apple Fruit
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

European Apple and Pear Crop Forecast 2011
EUROPEAN APPLE AND PEAR CROP FORECAST AUGUST 2011 9 10 August 2011 FOREWORD WAPA, the World Apple and Pear Association, is pleased to provide the 2011 European apple and pear crop estimate. This data will be released on the occasion of the 35th Prognosfruit Conference, which will take place in Ljubljana, Slovenia from 4-6 August 2011. This report is compiled upon the initiative of the apple and pear Working Group of COPA COGECA. The data has been collected with the useful support of the respective representative national producer organisations of the various Member States of the European Union and beyond. In regard to 2011, this report concludes that apple production in the EU for the 21 top producing countries contributing to this report will increase by 5% compared to the previous year, corresponding to a production of 10.195.000T. This production is however a 5% lower than the average for the last three years. In regard to varieties, Golden Delicious production will be up by 5% to 2.533.000T. Gala will increase by 7% to 1.059.000T. Jonagold will be up by 14% at 594.000T, while Red Delicious will decrease by 4% to 635.000T. Regarding pears, European growers predict a higher crop by 12% compared to 2010. Indeed, it is reported that the total crop in 2011 will reach 2.533.000T, compared to 2010 production which reached 2.264.000T. This figure relates to the production of the top 18 Member States of the EU-27 growing pears and contributing with their data to this report. -

Ardrossan Sets Sights on New Growth CONFIDENCE GROWS HERE
Australian Fruitgrower Winter 2020 • Vol 14 • Issue 2 Global knowledge at your fingertips SWP redefines ‘unskilled’ Pruning for vigour Ardrossan sets sights on new growth CONFIDENCE GROWS HERE Introducing a new level of confi dence in DMIs New Belanty fungicide gives you a new level of confi dence in the control of black spot in apples. Setting a new global standard for DMI’s, Belanty provides up to 100 times stronger enzyme binding than other DMI’s and is able to control resistant target disease populations. After years of research, Belanty is the breakthrough you’ve been waiting for. Find out more at crop-solutions.basf.com.au ALWAYS READ AND FOLLOW LABEL DIRECTIONS. © Copyright BASF 2020 ® Registered trademark of BASF. W244376 05.2020 CONTENTS A P A L NEWS LABOUR CEO Report . .04 Advocacy update . .05 Global knowledge at your fingertips . .12 21 FEA TURE 06 Seasonal workers redefine ‘unskilled’ Emerging BIOSECURITY stronger Biosecurity – what’s in it for me? . .26 Ardrossan sets sights on new growth . 06 RAISING T H E BAR : Tree returns must justify water cost . .08 R&D - LED INSIGH T S I N T O S M A R TER GROW T H Protect market access . .09 Focus on output . .09 Water security must come first . .10 Identify the opportunities . .11 28 EXPORT Online export training from July . .14 Pruning for vigour S T A T E R O UNDUP management State roundups . .16 MARKETING Campaign adds ‘feel good’ factor . .23 Pears that will arrive well before the heirs . 34 Sundial Orchard: Illuminating the future . -

Studies on Some Apple Virus Diseases in New Hampshire Joseph G
University of New Hampshire University of New Hampshire Scholars' Repository Doctoral Dissertations Student Scholarship Spring 1958 STUDIES ON SOME APPLE VIRUS DISEASES IN NEW HAMPSHIRE JOSEPH G. BARRAT Follow this and additional works at: https://scholars.unh.edu/dissertation Recommended Citation BARRAT, JOSEPH G., "STUDIES ON SOME APPLE VIRUS DISEASES IN NEW HAMPSHIRE" (1958). Doctoral Dissertations. 752. https://scholars.unh.edu/dissertation/752 This Dissertation is brought to you for free and open access by the Student Scholarship at University of New Hampshire Scholars' Repository. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of University of New Hampshire Scholars' Repository. For more information, please contact [email protected]. Dapple apple symptoms on the fruits of the variety Starking. STUDIES ON SOME APPLE VIRUS DISEASES IN NEW HAMPSHIRE By Joseph G. Barrat B. S., Rhode Island State College, 19*+8 M. S., University of Rhode Island, 1951 A DISSERTATION Submitted to the University of New Hampshire In Partial Fulfillment of The Requirements for the Degree of Doctor of Philosophy Graduate School Department of Botany May, 1958 This dissertation has been examined and approved. ■?. / T Date ACKNOWLEDGMENTS The writer wishes to express his deep appreciation to Dr. Avery E. Rich for his assistance and permission to develop the study along those lines which seemed most opportune. The writer is indebted to Dr. Albion R. Hodgdon for his taxonomic assistance, Dr. Stuart Dunn for permis sion to use the available space in the light room, Dr, R. A. Kilpatrick for help with the photographs and Dr. W. -

INF03 Reduce Lists of Apple Varieites
ECE/TRADE/C/WP.7/GE.1/2009/INF.3 Specialized Section on Standardization of Fresh Fruit and Vegetables Fifty-fifth session Geneva, 4 - 8 May 2009 Items 4(a) of the provisional agenda REVISION OF UNECE STANDARDS Proposals on the list of apple varieties This note has been put together by the secretariat following the decision taken by the Specialized Section at its fifty-fourth session to collect information from countries on varieties that are important in international trade. Replies have been received from the following countries: Canada, Czech Republic, Finland, France, Germany, Italy, Netherlands, New Zealand, Poland, Slovakia, South Africa, Sweden, Switzerland and the USA. This note also includes the documents compiled for the same purpose and submitted to the fifty-second session of the Specialized Section. I. Documents submitted to the 52nd session of the Specialized Section A. UNECE Standard for Apples – List of Varieties At the last meeting the 51 st session of the Specialized Section GE.1 the delegation of the United Kingdom offered to coordinate efforts to simplify the list of apple varieties. The aim was to see what the result would be if we only include the most important varieties that are produced and traded. The list is designed to help distinguish apple varieties by colour groups, size and russeting it is not exhaustive, non-listed varieties can still be marketed. The idea should not be to list every variety grown in every country. The UK asked for views on what were considered to be the most important top thirty varieties. Eight countries sent their views, Italy, Spain, the Netherlands, USA, Slovakia, Germany Finland and the Czech Republic. -

Rapid Pest Risk Analysis (PRA) For: Apple Dimple Fruit Viroid
Rapid Pest Risk Analysis (PRA) for: Apple dimple fruit viroid November 2018 1 Summary and conclusions of the rapid PRA Apple dimple fruit viroid (ADFVd) has the potential to make any infected apple crop unmarketable. However, since it was first identified in 1996, the viroid has only been identified in 5 different countries, and has a European presence in Italy only. The viroid does not appear to be spreading rapidly through trade or through natural means, and there is no evidence of a vector. As Malus species are widely distributed throughout the UK, it is thought that the viroid has the potential to establish in the UK. As the symptoms are very noticeable, it is thought that an outbreak would be detected rapidly with opportunities for containment and eradication. However, rapid detection could be confounded by some apple cultivars that are known to remain asymptomatic. This PRA shows that ADFVd is not a significant threat to apple production in the UK. Risk of entry The pathway of entry considered most likely is plants for planting of Malus species from Europe. Intra-EU trade of Malus species does exist, although there is no available data on the number of plants being traded. It is assumed that trade between Italy, where the viroid is present in the Campania region, and the UK does exist, and no mitigations to prevent entry of infected material are in place. As symptoms of ADFVd are only discernible on the fruit, it is impossible to detect the viroid in young plants without a molecular test. Plants for planting as a route for entry has been assessed as moderately likely with medium confidence. -

European Apple and Pear Crop Forecast 2012
EU 27 Apple Production by country x 1000 tons Country 2003 2004 2005 2006 2007 2008 2009 2010 2011 F2012 (1) (2) Austria 153 164 177 163 193 159 185 169 199 153 -23 -17 Belgium 320 356 317 358 358 336 344 288 305 212 -30 -32 Czech Rep 154 164 138 160 113 157 145 103 79 92 16 -16 Denmark 25 26 26 27 32 26 24 21 20 18 -10 -17 France 1.728 1.709 1.770 1.585 1.676 1.528 1.651 1.579 1.701 1.151 -32 -30 Germany 818 946 925 948 1.070 1.047 1.071 835 953 933 -2 -2 Greece 166 282 265 267 236 231 224 254 305 297 -3 14 Hungary 500 679 467 480 203 583 514 488 301 418 39 -4 Italy 2.144 2.032 2.085 1.991 2.196 2.164 2.237 2.179 2.293 1.993 -13 -11 Latvia 36 7 38 32 31 34 13 12 8 7 -13 -36 Lithuania 97 34 130 100 40 74 74 46 49 39 -20 -31 Netherlands 405 435 380 348 396 376 402 334 418 320 -23 -17 Poland 2.428 2.522 2.200 2.250 1.100 3.200 2.600 1.850 2.500 2.800 12 21 Portugal 287 277 252 257 258 245 274 251 265 225 -15 -15 Romania 811 1.098 439 417 287 459 379 423 412 402 -2 -1 Slovakia 34 31 36 31 10 42 48 32 33 39 18 4 Slovenia 62 62 58 71 80 68 64 66 73 51 -30 -25 Spain 704 553 701 547 599 528 470 486 507 384 -24 -21 Sweden 19 20 21 20 16 18 18 20 17 14 -18 -24 UK 157 164 193 174 196 201 212 214 226 191 -15 -12 Total: 11.048 11.560 10.617 10.226 9.091 11.476 10.948 9.651 10.663 9.739 -9 -7 (1) Percentage difference between F2012 and 2011 (2) Percentage difference between F2012 and the average of 2009 - 2010 - 2011 EU Apple Production 12000 10000 8000 6000 4000 2000 0 2003 2004 2005 2006 2007 2008 2009 2010 2011 F2012 EU 27 Apple Production by variety -

A Healthy Balance of Plasma Cholesterol by a Novel Annurca Apple-Based Nutraceutical Formulation: Results of a Randomized Trial
JOURNAL OF MEDICINAL FOOD J Med Food 00 (0) 2017, 1–13 FULL COMMUNICATION Mary Ann Liebert, Inc., and Korean Society of Food Science and Nutrition DOI: 10.1089/jmf.2016.0152 A Healthy Balance of Plasma Cholesterol by a Novel Annurca Apple-Based Nutraceutical Formulation: Results of a Randomized Trial Gian Carlo Tenore,1 Domenico Caruso,2 Giuseppe Buonomo,3 Maria D’Avino,2 Pietro Campiglia,4 Luciana Marinelli,1 and Ettore Novellino1 1Department of Pharmacy, University of Naples Federico II, Naples, Italy. 2Department of Internal Medicine, Hospital Cardarelli, Naples, Italy. 3Coop. Samnium Medica, Benevento, Italy. 4Department of Pharmaceutical and Biomedical Sciences, University of Salerno, Salerno, Italy. ABSTRACT Cardiovascular diseases are nowadays preferential targets of preventive medicine through a straightforward therapy on lipid profile. However, statins, the first-line lipid-lowering drug therapy, specifically act on low-density lipoprotein cholesterol (LDL-C), having a modest effect on plasma high-density lipoprotein cholesterol (HDL-C) concentrations. Today, a number of novel HDL-targeted therapies are emerging, along with unexpected side effects. Thus, novel and possibly safe substances, able to correct impaired lipid profile in humans, are still in great demand. Herein, based on encouraging clinical data, we formulated a nutraceutical product (AppleMetSÒ, AMS), based on a polyphenolic extract from Annurca apple, and demonstrated that two capsules a day of AMS, after one month, have a LDL-C lowering outcome equivalent to 40 mg of simvastatin or 10 mg of atorvastatin. Nevertheless, different from statin-based therapy, AMS exerted a notable effect on HDL (+49.2%). Based on the trial results, we can assert that AMS formulation could effectively integrate the current therapeutic arsenal to correct impaired lipid profile in humans. -

EFFECT of WATER REGIMES on YIELD and QUALITY of APPLE UNDER SEMI-ARID, LIBYA Ahmed M
Misr J. Ag. Eng., 30 (2): 327 - 346 IRRIGATION AND DRAINAGE EFFECT OF WATER REGIMES ON YIELD AND QUALITY OF APPLE UNDER SEMI-ARID, LIBYA Ahmed M. Hassan* ABSTRACT This study, was carried out in 2011/2012, to investigate the yield and quality parameters of apple fruits (double red delicious) with different irrigation systems; surface and trickle irrigation and with different water levels; 100, 75 and 50 % of ETc. Traditional irrigation (rainfall irrigation) presented 34.8% from ETc. The water irrigation requirement was determined by using Penman-Monteith’s equation. An increase in fruit yield, properties and quality (fruit diameter, fruit weight, total soluble solid and sugar) has been noted with trickle irrigation method comparing with surface and rainfall irrigations. The highest fruit diameter (80.9 mm), fruit mass (216.3 g), extra and class 1 fruit ratios (36.2 and 36.5%), total soluble solid (1.551 ton/fed) and sugar (1.38 ton/fed) were observed with DET100 treatment. To obtain a high quantity and quality apples, DET100 treatment with 100% ETc and trickle irrigation system are recommended during transition from rainfall and surface irrigation to trickle irrigation for similar climatic and soil conditions. The results showed that trickle irrigation system increases the qualitative and quantitative properties of apple fruits. Key words: surface irrigation, trickle irrigation, deficit irrigation, apple, yield, water use efficiency, quality parameters. INTRODUCTION eficit irrigation is a strategy which allows a crop to sustain some degree of water deficit in order to reduce irrigation costs and D potentially increase revenues. English and Raja (1996) described three deficit irrigation case studies in which the reductions in irrigation costs were greater than the reductions in revenue due to reduced yields. -

R Graphics Output
Aberystwyth University Development of a minimal KASP marker panel for distinguishing genotypes in apple collections Winfield, Mark; Burridge, Amanda; Ordidge, Matthew; Harper, Helen; Wilkinson, Paul; Thorogood, Danny; Copas, Liz; Edwards, Keith; Barker, Gary Published in: PLoS One DOI: 10.1371/journal.pone.0242940 Publication date: 2020 Citation for published version (APA): Winfield, M., Burridge, A., Ordidge, M., Harper, H., Wilkinson, P., Thorogood, D., Copas, L., Edwards, K., & Barker, G. (2020). Development of a minimal KASP marker panel for distinguishing genotypes in apple collections. PLoS One, 15(11), [e0242940]. https://doi.org/10.1371/journal.pone.0242940 Document License CC BY General rights Copyright and moral rights for the publications made accessible in the Aberystwyth Research Portal (the Institutional Repository) are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the Aberystwyth Research Portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the Aberystwyth Research Portal Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. -
MELO Seconda Parte.Pdf
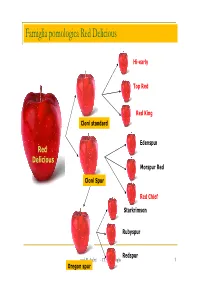
Famiglia pomologica Red Delicious Hi-early Top Red Red King Cloni standard Edenspur Red Delicious Morspur Red Cloni Spur Red Chief Starkrimson Rubyspur Redspur prof. R. Andrei - I.T.A.S- Treviglio 1 Oregon spur Fuji Giapponese, di recente introduzione in Europa Pianta molto vigorosa adatta alle forme libere, con buona produzione e precoce entrata in produzione Frutto di forma cilindrica, a polpa croccante, soda molto zuccherina e profumata Buccia rosso-rosato Molto produttiva, necessita del diradamento Raccolta II decade di ottobre prof. R. Andrei - I.T.A.S- Treviglio 2 Famiglia pomologica Fuji Fuji kiku 8 Fuji Fuji naga-fu 12 prof. R. Andrei - I.T.A.S- Treviglio 3 Jonagold Varietà originata nel 1943 negli Stati Uniti, da un incrocio di Golden Delicious e Jonathan Diffusa in Europa dalla fine degli anni sessanta, è oggi una delle varietà più conosciute e apprezzate specialmente nel centro e nord Europa Frutto grosso, rotondo, presenta una colorazione di fondo verde- gialla e copertura di rosso carminio. Buccia liscia, leggermente cerosa Delicatamente croccante e con la polpa color crema, presenta un ottimo aroma dolce aromatico prof. R. Andrei - I.T.A.S- Treviglio 4 Famiglia pomologica Jonagold Renetta del Canada Jonagold Querina Novajo prof. R. Andrei - I.T.A.S- Treviglio 5 Pink Lady Nata da un'incrocio tra Williams e Golden Delicious, Cripps Pink è stata creata alla stazione di ricerche di Stoneville (Australia occidentale) nel 1973. E' presente sui mercati solo con il nome Pink Lady ®, che è il marchio commerciale della varietà. La forma del frutto è da oblunga a cilindrica ed omogenea. -

Formulation of Novel Functional Foods and Nutraceutical Products from Annurca Apple with Healthy Human Effects
UNIVERSITY OF NAPLES – FEDERICO II DEPARTMENT OF PHARMACY Ph.D. THESIS IN “PHARMACEUTICAL SCIENCE” Formulation of novel Functional Foods and Nutraceutical Products from Annurca apple with healthy human effects Supervisor Coordinator Prof. Alberto Ritieni Prof. Maria Valeria D’Auria Candidate Dr. Maria Maisto XXX CYCLE Abstract 1 Research objectives 2 Introduction 3 Annurca apple the queen of the apples: an overview of history, origin and cultivation 6 Polyphenols 8 Human biological activity 14 Bioaccessibility and bioavailability 16 Polyphenols contained in apples 18 Chapter 1 Functional Food formulation Annurca apple puree enriched with different strains of acid lactic bacteria as a potential functional food: In vitro and in vivo studies 1.1 Introduction 23 1.2 Matherials and Methods 25 1.2.1 Apple collection 26 1.2.2 Annurca apple puree preparation 26 1.2.3 Fermentation Annurca apple puree 26 1.2.4 pH measurment 26 1.2.5 Biochemical analysis 26 1.2.6 Antioxidant activity 26 1.2.7 Total phenol content 27 1.2.8 Study population and protocol 27 1.2.9 Randomisation, concealment and blinding 28 1.2.10 Study treatments 28 1.2.11 Study outcomes and data colection 30 1.2.12 Safety 30 1.2.13 TMAO LC-MS Analysis 30 1.2.14 Statistical analysis 30 i 1.3 Results 31 1.3.1 pH reductions 31 1.3.2 Antioxidant activity and total phenol content of fermented products 33 1.3.3 Blood TMAO concentration 35 1.4 Discussion 35 1.5 Conclusion 37 Chapter 2 Nutraceutical Formulations 2.1 Annurca apple nutraceutical formulation enhances keratin expression in a human -

Identification of Selective Sweeps in Domesticated
Sede Amministrativa: Universita` degli studi di Padova Dipartimento di Biologia SCUOLA DI DOTTORATO DI RICERCA IN: Bioscienze e Biotecnologie INDIRIZZO: Biotecnologie CICLO: XXVII Identification of selective sweeps in domesticated apple (Malus × domestica Borkh.) Direttore della Scuola: Ch.mo Prof. Giuseppe Zanotti Coordinatore d'indirizzo: Ch.mo Prof. Fiorella Lo Schiavo Supervisore: Ch.mo Prof. Gorgio Valle Dottorando : Emanuela Kerschbamer The apple is so deeply rooted in the culture of human populations from temperate regions that it is often not recognized as an exotic plant of unclear origin. Cornille et al. 2012 iv Contents 1 Riassunto 1 2 Abstract 3 3 Introduction 5 3.1 Apple . .5 3.2 Plant domestication . .8 3.2.1 Apple domestication . .9 3.3 Genetic diversity . 11 3.3.1 Single Nucleotide Polymorphism . 12 3.3.2 Linkage disequilibrium . 13 3.4 Selection . 15 3.4.1 Selective sweeps . 16 4 Aims of the project 21 5 Materials and methods 23 5.1 Samples and data . 23 5.2 Alignment and SNP calling . 25 5.3 SNP filtering . 26 5.4 Population structure . 27 5.5 Population genetic analyses . 28 5.6 Identification of regions under selection . 30 5.7 Candidate genes annotation . 31 i CONTENTS 6 Results 33 6.1 Alignment, SNP calling and filtering . 33 6.2 Population structure . 36 6.3 Genetic variability . 39 6.4 Selective sweep analysis . 40 6.5 Annotation . 43 7 Discussion 59 7.1 SNP calling and filtering . 59 7.2 Population structure . 60 7.3 Genetic variability . 61 7.4 Selective sweep analysis . 62 8 Conclusion 69 9 Appendix A: Apple Cultivars 71 9.1 ....................................