Electrophoresis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cyclin D1/Cyclin-Dependent Kinase 4 Interacts with Filamin a and Affects the Migration and Invasion Potential of Breast Cancer Cells
Published OnlineFirst February 28, 2010; DOI: 10.1158/0008-5472.CAN-08-1108 Tumor and Stem Cell Biology Cancer Research Cyclin D1/Cyclin-Dependent Kinase 4 Interacts with Filamin A and Affects the Migration and Invasion Potential of Breast Cancer Cells Zhijiu Zhong, Wen-Shuz Yeow, Chunhua Zou, Richard Wassell, Chenguang Wang, Richard G. Pestell, Judy N. Quong, and Andrew A. Quong Abstract Cyclin D1 belongs to a family of proteins that regulate progression through the G1-S phase of the cell cycle by binding to cyclin-dependent kinase (cdk)-4 to phosphorylate the retinoblastoma protein and release E2F transcription factors for progression through cell cycle. Several cancers, including breast, colon, and prostate, overexpress the cyclin D1 gene. However, the correlation of cyclin D1 overexpression with E2F target gene regulation or of cdk-dependent cyclin D1 activity with tumor development has not been identified. This suggests that the role of cyclin D1 in oncogenesis may be independent of its function as a cell cycle regulator. One such function is the role of cyclin D1 in cell adhesion and motility. Filamin A (FLNa), a member of the actin-binding filamin protein family, regulates signaling events involved in cell motility and invasion. FLNa has also been associated with a variety of cancers including lung cancer, prostate cancer, melanoma, human bladder cancer, and neuroblastoma. We hypothesized that elevated cyclin D1 facilitates motility in the invasive MDA-MB-231 breast cancer cell line. We show that MDA-MB-231 motility is affected by disturbing cyclin D1 levels or cyclin D1-cdk4/6 kinase activity. -

Genetic Mutations and Mechanisms in Dilated Cardiomyopathy
Genetic mutations and mechanisms in dilated cardiomyopathy Elizabeth M. McNally, … , Jessica R. Golbus, Megan J. Puckelwartz J Clin Invest. 2013;123(1):19-26. https://doi.org/10.1172/JCI62862. Review Series Genetic mutations account for a significant percentage of cardiomyopathies, which are a leading cause of congestive heart failure. In hypertrophic cardiomyopathy (HCM), cardiac output is limited by the thickened myocardium through impaired filling and outflow. Mutations in the genes encoding the thick filament components myosin heavy chain and myosin binding protein C (MYH7 and MYBPC3) together explain 75% of inherited HCMs, leading to the observation that HCM is a disease of the sarcomere. Many mutations are “private” or rare variants, often unique to families. In contrast, dilated cardiomyopathy (DCM) is far more genetically heterogeneous, with mutations in genes encoding cytoskeletal, nucleoskeletal, mitochondrial, and calcium-handling proteins. DCM is characterized by enlarged ventricular dimensions and impaired systolic and diastolic function. Private mutations account for most DCMs, with few hotspots or recurring mutations. More than 50 single genes are linked to inherited DCM, including many genes that also link to HCM. Relatively few clinical clues guide the diagnosis of inherited DCM, but emerging evidence supports the use of genetic testing to identify those patients at risk for faster disease progression, congestive heart failure, and arrhythmia. Find the latest version: https://jci.me/62862/pdf Review series Genetic mutations and mechanisms in dilated cardiomyopathy Elizabeth M. McNally, Jessica R. Golbus, and Megan J. Puckelwartz Department of Human Genetics, University of Chicago, Chicago, Illinois, USA. Genetic mutations account for a significant percentage of cardiomyopathies, which are a leading cause of conges- tive heart failure. -

Plakoglobin Is Required for Effective Intermediate Filament Anchorage to Desmosomes Devrim Acehan1, Christopher Petzold1, Iwona Gumper2, David D
ORIGINAL ARTICLE Plakoglobin Is Required for Effective Intermediate Filament Anchorage to Desmosomes Devrim Acehan1, Christopher Petzold1, Iwona Gumper2, David D. Sabatini2, Eliane J. Mu¨ller3, Pamela Cowin2,4 and David L. Stokes1,2,5 Desmosomes are adhesive junctions that provide mechanical coupling between cells. Plakoglobin (PG) is a major component of the intracellular plaque that serves to connect transmembrane elements to the cytoskeleton. We have used electron tomography and immunolabeling to investigate the consequences of PG knockout on the molecular architecture of the intracellular plaque in cultured keratinocytes. Although knockout keratinocytes form substantial numbers of desmosome-like junctions and have a relatively normal intercellular distribution of desmosomal cadherins, their cytoplasmic plaques are sparse and anchoring of intermediate filaments is defective. In the knockout, b-catenin appears to substitute for PG in the clustering of cadherins, but is unable to recruit normal levels of plakophilin-1 and desmoplakin to the plaque. By comparing tomograms of wild type and knockout desmosomes, we have assigned particular densities to desmoplakin and described their interaction with intermediate filaments. Desmoplakin molecules are more extended in wild type than knockout desmosomes, as if intermediate filament connections produced tension within the plaque. On the basis of our observations, we propose a particular assembly sequence, beginning with cadherin clustering within the plasma membrane, followed by recruitment of plakophilin and desmoplakin to the plaque, and ending with anchoring of intermediate filaments, which represents the key to adhesive strength. Journal of Investigative Dermatology (2008) 128, 2665–2675; doi:10.1038/jid.2008.141; published online 22 May 2008 INTRODUCTION dense plaque that is further from the membrane and that Desmosomes are large macromolecular complexes that mediates the binding of intermediate filaments. -

Molecular Pathways Associated with the Nutritional Programming of Plant-Based Diet Acceptance in Rainbow Trout Following an Early Feeding Exposure Mukundh N
Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, Aurélie Le Cam, Françoise Médale, Sadasivam J. Kaushik, Inge Geurden To cite this version: Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, et al.. Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure. BMC Genomics, BioMed Central, 2016, 17 (1), pp.1-20. 10.1186/s12864-016-2804-1. hal-01346912 HAL Id: hal-01346912 https://hal.archives-ouvertes.fr/hal-01346912 Submitted on 19 Jul 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Balasubramanian et al. BMC Genomics (2016) 17:449 DOI 10.1186/s12864-016-2804-1 RESEARCH ARTICLE Open Access Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian1, Stephane Panserat1, Mathilde Dupont-Nivet2, Edwige Quillet2, Jerome Montfort3, Aurelie Le Cam3, Francoise Medale1, Sadasivam J. Kaushik1 and Inge Geurden1* Abstract Background: The achievement of sustainable feeding practices in aquaculture by reducing the reliance on wild-captured fish, via replacement of fish-based feed with plant-based feed, is impeded by the poor growth response seen in fish fed high levels of plant ingredients. -
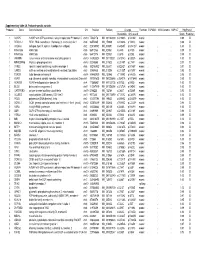
Supplementary Table 2A. Proband-Specific Variants Proband
Supplementary table 2A. Proband-specific variants Proband Gene Gene full name Chr Position RefSeqChange Function ESP6500 1000 Genome SNP137 PolyPhen2 Nucleotide Amino acid Score Prediction 1 AGAP5 ArfGAP with GTPase domain, ankyrin repeat and PH domain 5 chr10 75435178 NM_001144000 c.G1240A p.V414M exonic . 0.99 D 1 REXO1L1 REX1, RNA exonuclease 1 homolog (S. cerevisiae)-like 1 chr8 86574465 NM_172239 c.A1262G p.Y421C exonic . 0.98 D 1 COL4A3 collagen, type IV, alpha 3 (Goodpasture antigen) chr2 228169782 NM_000091 c.G4235T p.G1412V exonic . 1.00 D 1 KIAA1586 KIAA1586 chr6 56912133 NM_020931 c.C44G p.A15G exonic . 0.99 D 1 KIAA1586 KIAA1586 chr6 56912176 NM_020931 c.C87G p.D29E exonic . 0.99 D 1 JAKMIP3 Janus kinase and microtubule interacting protein 3 chr10 133955524 NM_001105521 c.G1574C p.G525A exonic . 1.00 D 1 MPHOSPH8 M-phase phosphoprotein 8 chr13 20240685 NM_017520 c.C2140T p.L714F exonic . 0.97 D 1 SYNE1 spectrin repeat containing, nuclear envelope 1 chr6 152783902 NM_033071 c.A2242T p.N748Y exonic . 0.97 D 1 CAND2 cullin-associated and neddylation-dissociated 2 (putative) chr3 12858835 NM_012298 c.C2125T p.H709Y exonic . 0.95 D 1 TDRD9 tudor domain containing 9 chr14 104460909 NM_153046 c.T1289G p.V430G exonic . 0.96 D 2 ASPM asp (abnormal spindle) homolog, microcephaly associated (Drosophila)chr1 197091625 NM_001206846 c.G3491A p.R1164H exonic . 0.99 D 2 ADAM29 ADAM metallopeptidase domain 29 chr4 175896951 NM_001130705 c.A275G p.D92G exonic . 1.00 D 2 BCO2 beta-carotene oxygenase 2 chr11 112087019 NM_001256398 c.G1373A p.G458E exonic . -

Plakophilin-2 Haploinsufficiency Causes Calcium Handling
International Journal of Molecular Sciences Article Plakophilin-2 Haploinsufficiency Causes Calcium Handling Deficits and Modulates the Cardiac Response Towards Stress Chantal J.M. van Opbergen 1 , Maartje Noorman 1, Anna Pfenniger 2, Jaël S. Copier 1, Sarah H. Vermij 2,3 , Zhen Li 2, Roel van der Nagel 1, Mingliang Zhang 2, Jacques M.T. de Bakker 1,4, Aaron M. Glass 5, Peter J. Mohler 6,7, Steven M. Taffet 5, Marc A. Vos 1, Harold V.M. van Rijen 1, Mario Delmar 2 and Toon A.B. van Veen 1,* 1 Department of Medical Physiology, Division of Heart & Lungs, University Medical Center Utrecht, Yalelaan 50, 3584CM Utrecht, The Netherlands 2 Division of Cardiology, NYU School of Medicine, New York, NY 10016, USA 3 Institute of Biochemistry and Molecular Medicine, University of Bern, 3012 Bern, Switzerland 4 Department of Medical Biology, Academic Medical Center Amsterdam, 1105AZ Amsterdam, The Netherlands 5 Department of Microbiology and Immunology, SUNY Upstate Medical University, Syracuse, NY 13210, USA 6 Dorothy M. Davis Heart and Lung Research Institute, The Ohio State University College of Medicine and Wexner Medical Center, Columbus, OH 43210, USA 7 Departments of Physiology & Cell Biology and Internal Medicine, Division of Cardiovascular Medicine, The Ohio State University College of Medicine Wexner Medical Center, Columbus, OH 43210, USA * Correspondence: [email protected] Received: 1 August 2019; Accepted: 19 August 2019; Published: 21 August 2019 Abstract: Human variants in plakophilin-2 (PKP2) associate with most cases of familial arrhythmogenic cardiomyopathy (ACM). Recent studies show that PKP2 not only maintains intercellular coupling, but also regulates transcription of genes involved in Ca2+ cycling and cardiac rhythm. -

The Ubiquitin-Proteasome Pathway Mediates Gelsolin Protein Downregulation in Pancreatic Cancer
The Ubiquitin-Proteasome Pathway Mediates Gelsolin Protein Downregulation in Pancreatic Cancer Xiao-Guang Ni,1 Lu Zhou,2 Gui-Qi Wang,1 Shang-Mei Liu,3 Xiao-Feng Bai,4 Fang Liu,5 Maikel P Peppelenbosch,2 and Ping Zhao4 1Department of Endoscopy, Cancer Institute and Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China; 2Department of Cell Biology, University Medical Center Groningen, University of Groningen, Groningen, The Netherlands; 3Department of Pathology, 4Department of Abdominal Surgery, and 5State Key Laboratory of Molecular Oncology, Cancer Institute and Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China A well-known observation with respect to cancer biology is that transformed cells display a disturbed cytoskeleton. The under- lying mechanisms, however, remain only partly understood. In an effort to identify possible mechanisms, we compared the pro- teome of pancreatic cancer with matched normal pancreas and observed diminished protein levels of gelsolin—an actin fila- ment severing and capping protein of crucial importance for maintaining cytoskeletal integrity—in pancreatic cancer. Additionally, pancreatic ductal adenocarcinomas displayed substantially decreased levels of gelsolin as judged by Western blot and immunohistochemical analyses of tissue micoarrays, when compared with cancerous and untransformed tissue from the same patients (P < 0.05). Importantly, no marked downregulation of gelsolin mRNA was observed (P > 0.05), suggesting that post- transcriptional mechanisms mediate low gelsolin protein levels. In apparent agreement, high activity ubiquitin-proteasome path- way in both patient samples and the BxPC-3 pancreatic cancer cell line was detected, and inhibition of the 26s proteasome sys- tem quickly restored gelsolin protein levels in the latter cell line. -

Structural and Biochemical Changes Underlying a Keratoderma-Like Phenotype in Mice Lacking Suprabasal AP1 Transcription Factor Function
Citation: Cell Death and Disease (2015) 6, e1647; doi:10.1038/cddis.2015.21 OPEN & 2015 Macmillan Publishers Limited All rights reserved 2041-4889/15 www.nature.com/cddis Structural and biochemical changes underlying a keratoderma-like phenotype in mice lacking suprabasal AP1 transcription factor function EA Rorke*,1, G Adhikary2, CA Young2, RH Rice3, PM Elias4, D Crumrine4, J Meyer4, M Blumenberg5 and RL Eckert2,6,7,8 Epidermal keratinocyte differentiation on the body surface is a carefully choreographed process that leads to assembly of a barrier that is essential for life. Perturbation of keratinocyte differentiation leads to disease. Activator protein 1 (AP1) transcription factors are key controllers of this process. We have shown that inhibiting AP1 transcription factor activity in the suprabasal murine epidermis, by expression of dominant-negative c-jun (TAM67), produces a phenotype type that resembles human keratoderma. However, little is understood regarding the structural and molecular changes that drive this phenotype. In the present study we show that TAM67-positive epidermis displays altered cornified envelope, filaggrin-type keratohyalin granule, keratin filament, desmosome formation and lamellar body secretion leading to reduced barrier integrity. To understand the molecular changes underlying this process, we performed proteomic and RNA array analysis. Proteomic study of the corneocyte cross-linked proteome reveals a reduction in incorporation of cutaneous keratins, filaggrin, filaggrin2, late cornified envelope precursor proteins, hair keratins and hair keratin-associated proteins. This is coupled with increased incorporation of desmosome linker, small proline-rich, S100, transglutaminase and inflammation-associated proteins. Incorporation of most cutaneous keratins (Krt1, Krt5 and Krt10) is reduced, but incorporation of hyperproliferation-associated epidermal keratins (Krt6a, Krt6b and Krt16) is increased. -

CDH12 Cadherin 12, Type 2 N-Cadherin 2 RPL5 Ribosomal
5 6 6 5 . 4 2 1 1 1 2 4 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 A A A A A A A A A A A A A A A A A A A A C C C C C C C C C C C C C C C C C C C C R R R R R R R R R R R R R R R R R R R R B , B B B B B B B B B B B B B B B B B B B , 9 , , , , 4 , , 3 0 , , , , , , , , 6 2 , , 5 , 0 8 6 4 , 7 5 7 0 2 8 9 1 3 3 3 1 1 7 5 0 4 1 4 0 7 1 0 2 0 6 7 8 0 2 5 7 8 0 3 8 5 4 9 0 1 0 8 8 3 5 6 7 4 7 9 5 2 1 1 8 2 2 1 7 9 6 2 1 7 1 1 0 4 5 3 5 8 9 1 0 0 4 2 5 0 8 1 4 1 6 9 0 0 6 3 6 9 1 0 9 0 3 8 1 3 5 6 3 6 0 4 2 6 1 0 1 2 1 9 9 7 9 5 7 1 5 8 9 8 8 2 1 9 9 1 1 1 9 6 9 8 9 7 8 4 5 8 8 6 4 8 1 1 2 8 6 2 7 9 8 3 5 4 3 2 1 7 9 5 3 1 3 2 1 2 9 5 1 1 1 1 1 1 5 9 5 3 2 6 3 4 1 3 1 1 4 1 4 1 7 1 3 4 3 2 7 6 4 2 7 2 1 2 1 5 1 6 3 5 6 1 3 6 4 7 1 6 5 1 1 4 1 6 1 7 6 4 7 e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m -

140503 IPF Signatures Supplement Withfigs Thorax
Supplementary material for Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis Daryle J. DePianto1*, Sanjay Chandriani1⌘*, Alexander R. Abbas1, Guiquan Jia1, Elsa N. N’Diaye1, Patrick Caplazi1, Steven E. Kauder1, Sabyasachi Biswas1, Satyajit K. Karnik1#, Connie Ha1, Zora Modrusan1, Michael A. Matthay2, Jasleen Kukreja3, Harold R. Collard2, Jackson G. Egen1, Paul J. Wolters2§, and Joseph R. Arron1§ 1Genentech Research and Early Development, South San Francisco, CA 2Department of Medicine, University of California, San Francisco, CA 3Department of Surgery, University of California, San Francisco, CA ⌘Current address: Novartis Institutes for Biomedical Research, Emeryville, CA. #Current address: Gilead Sciences, Foster City, CA. *DJD and SC contributed equally to this manuscript §PJW and JRA co-directed this project Address correspondence to Paul J. Wolters, MD University of California, San Francisco Department of Medicine Box 0111 San Francisco, CA 94143-0111 [email protected] or Joseph R. Arron, MD, PhD Genentech, Inc. MS 231C 1 DNA Way South San Francisco, CA 94080 [email protected] 1 METHODS Human lung tissue samples Tissues were obtained at UCSF from clinical samples from IPF patients at the time of biopsy or lung transplantation. All patients were seen at UCSF and the diagnosis of IPF was established through multidisciplinary review of clinical, radiological, and pathological data according to criteria established by the consensus classification of the American Thoracic Society (ATS) and European Respiratory Society (ERS), Japanese Respiratory Society (JRS), and the Latin American Thoracic Association (ALAT) (ref. 5 in main text). Non-diseased normal lung tissues were procured from lungs not used by the Northern California Transplant Donor Network. -

Alpha;-Actinin-4 Promotes Metastasis in Gastric Cancer
Laboratory Investigation (2017) 97, 1084–1094 © 2017 USCAP, Inc All rights reserved 0023-6837/17 α-Actinin-4 promotes metastasis in gastric cancer Xin Liu and Kent-Man Chu Metastasis increases the mortality rate of gastric cancer, which is the third leading cause of cancer-associated deaths worldwide. This study aims to identify the genes promoting metastasis of gastric cancer (GC). A human cell motility PCR array was used to analyze a pair of tumor and non-tumor tissue samples from a patient with stage IV GC (T3N3M1). Expression of the dysregulated genes was then evaluated in GC tissue samples (n = 10) and cell lines (n =6) via qPCR. Expression of α-actinin-4 (ACTN4) was validated in a larger sample size (n = 47) by qPCR, western blot and immunohistochemistry. Knockdown of ACTN4 with specific siRNAs was performed in GC cells, and adhesion assays, transwell invasion assays and migration assays were used to evaluate the function of these cells. Expression of potential targets of ACTN4 were then evaluated by qPCR. Thirty upregulated genes (greater than twofold) were revealed by the PCR array. We focused on ACTN4 because it was upregulated in 6 out of 10 pairs of tissue samples and 5 out of 6 GC cell lines. Further study indicated that ACTN4 was upregulated in 22/32 pairs of tissue samples at stage III & IV (P = 0.0069). Knockdown of ACTN4 in GC cells showed no significant effect on cell proliferation, but significantly increased cell-matrix adhesion, as well as reduced migration and invasion of AGS, MKN7 and NCI-N87 cells. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase