Phylogenies and Secondary Chemistry in Arnica (Asteraceae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

"National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary."
Intro 1996 National List of Vascular Plant Species That Occur in Wetlands The Fish and Wildlife Service has prepared a National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary (1996 National List). The 1996 National List is a draft revision of the National List of Plant Species That Occur in Wetlands: 1988 National Summary (Reed 1988) (1988 National List). The 1996 National List is provided to encourage additional public review and comments on the draft regional wetland indicator assignments. The 1996 National List reflects a significant amount of new information that has become available since 1988 on the wetland affinity of vascular plants. This new information has resulted from the extensive use of the 1988 National List in the field by individuals involved in wetland and other resource inventories, wetland identification and delineation, and wetland research. Interim Regional Interagency Review Panel (Regional Panel) changes in indicator status as well as additions and deletions to the 1988 National List were documented in Regional supplements. The National List was originally developed as an appendix to the Classification of Wetlands and Deepwater Habitats of the United States (Cowardin et al.1979) to aid in the consistent application of this classification system for wetlands in the field.. The 1996 National List also was developed to aid in determining the presence of hydrophytic vegetation in the Clean Water Act Section 404 wetland regulatory program and in the implementation of the swampbuster provisions of the Food Security Act. While not required by law or regulation, the Fish and Wildlife Service is making the 1996 National List available for review and comment. -

38 Plant Species at Risk Accounts in Prince George Timber Supply Area
38 PLANT SPECIES AT RISK ACCOUNTS IN PRINCE GEORGE TIMBER SUPPLY AREA FIA PROJECT 2668067 Prepared for: Canadian Forest Products Ltd. 5162 Northwood Pulpmill Road Prince George, B.C. V2L 4W2 Attn: Bruce Bradley Prepared by: Timberline Natural Resource Group Ltd. 1512 97 th Avenue Dawson Creek, BC V1G 1N7 March 2008 1 Acknowlegements The authors of these accounts, Mecah Klem, Jen Atkins, Korey Green and Dan Bernier would like to thank Bruce Bradley and the PG FIA licensee group for funding this project. We would also like to acknowledge the work by Timberline staff on related projects – their research and previous related technical reports were useful for completing these accounts in an efficient and comprehensive manner. Gilbert Proulx of Alpha Wildlife Research and Management is also thanked for his contribution to other components of this collaborative project. 2 Table of Contents: Introduction ................................................................................................... 5 Alpine cliff fern .............................................................................................. 6 American sweet-flag ...................................................................................... 8 Arctic rush ................................................................................................... 10 Austrian draba ............................................................................................ 12 Back’s sedge ................................................................................................ -

State of Colorado 2016 Wetland Plant List
5/12/16 State of Colorado 2016 Wetland Plant List Lichvar, R.W., D.L. Banks, W.N. Kirchner, and N.C. Melvin. 2016. The National Wetland Plant List: 2016 wetland ratings. Phytoneuron 2016-30: 1-17. Published 28 April 2016. ISSN 2153 733X http://wetland-plants.usace.army.mil/ Aquilegia caerulea James (Colorado Blue Columbine) Photo: William Gray List Counts: Wetland AW GP WMVC Total UPL 83 120 101 304 FACU 440 393 430 1263 FAC 333 292 355 980 FACW 342 329 333 1004 OBL 279 285 285 849 Rating 1477 1419 1504 1511 User Notes: 1) Plant species not listed are considered UPL for wetland delineation purposes. 2) A few UPL species are listed because they are rated FACU or wetter in at least one Corps Region. 3) Some state boundaries lie within two or more Corps Regions. If a species occurs in one region but not the other, its rating will be shown in one column and the other column will be BLANK. Approved for public release; distribution is unlimited. 1/22 5/12/16 Scientific Name Authorship AW GP WMVC Common Name Abies bifolia A. Murr. FACU FACU Rocky Mountain Alpine Fir Abutilon theophrasti Medik. UPL UPL FACU Velvetleaf Acalypha rhomboidea Raf. FACU FACU Common Three-Seed-Mercury Acer glabrum Torr. FAC FAC FACU Rocky Mountain Maple Acer grandidentatum Nutt. FACU FAC FACU Canyon Maple Acer negundo L. FACW FAC FAC Ash-Leaf Maple Acer platanoides L. UPL UPL FACU Norw ay Maple Acer saccharinum L. FAC FAC FAC Silver Maple Achillea millefolium L. FACU FACU FACU Common Yarrow Achillea ptarmica L. -

DECISION NOTICE/DESIGNATION ORDER Decision Notice Finding Of
DECISION NOTICE/DESIGNATION ORDER Decision Notice Finding of No Significant Impact Designation Order By virtue of the authority vested in me by the Secretary of Agriculture under regulations at 7 CFR 2.42, 36 CFR 251.23, and 36 CFR Part 219, I hereby establish the Osborn Mountain Research Natural Area. It shall be comprised of lands described in the section of the Establishment Record entitled "Location." The Regional Forester has recommended the establishment of this Research Natural Area in the Record of Decision for the Bridger-Teton National Forest Land and Resource Management Plan. That recommendation was the result of analysis of the factors listed in 36 CFR 219.25 and Forest Service Manual 4063.41. Results of the Regional Forester's analysis are documented in the Bridger-Teton National Forest Land and Resource Management Plan and Final Environmental Impact Statement which are available to the public. The Osborn Mountain Research Natural Area will be managed in compliance with all relevant laws, regulations, and Forest Service Manual direction regarding Research Natural Areas. It will be administered in accordance with the management direction/prescription identified in the Establishment Record. I have reviewed the Bridger-Teton Land and Resource Management Plan (LRMP) direction for this RNA and find that the management direction cited in the previous paragraph is consistent with the LRMP and that a Plan amendment is not required. The Forest Supervisor of the Bridger-Teton National Forest shall notify the public of this decision and mail a copy of the Decision Notice/Designation Order and amended direction to all persons on the Bridger-Teton National Forest Land and Resource Management Plan mailing list. -

Conservation of Eastern European Medicinal Plants Arnica Montana in Romania Management Plan
Conservation of Eastern European Medicinal Plants Arnica montana in Romania Case Study Gârda de Sus Management Plan Barbara Michler 2007 Projekt Leader: Dr. Susanne Schmitt, Dr. Wolfgang Kathe (maternity cover) WWF-UK Panda House, Weyside Park, Godalming, Surrey GU7 1XR, United Kingdom Administration: Michael Balzer and team WWF-DCP Mariahilfer-Str. 88a/3/9 A-1070 Wien Austria Projekt Manager: Maria Mihul WWF-DCP 61, Marastu Bdv. 3rd floor, 326/327/328 Sector 1, Bucharest, RO-71331 Romania With financial support of the Darwin Initiative Area 3D, Third Floor, Nobel House 17 Smith Square, London SW1P 3JR United Kingdom Project Officer: Dr. Barbara Michler Dr. Fischer, ifanos-Landschaftsökologie Forchheimer Weg 46 D-91341 Röttenbach Germany Local Coordinator: Dr. Florin Pacurar University of Agricultural Sciences and Veterinary Medicine (USAMV) Department of Fodder Production & Conservation Cluj-Napoca, Romania Major of the community Gârda de Sus Alba Iulia Romania Acknowledgements I am very grateful to a number of people who were involved in the process of the project over the last 6 years (including 3 previous years under Project Apuseni). Thanks to all of them (alphabetic order): Apuseni Nature Park: Alin Mos Arnica project team: Mona Cosma, Valentin Dumitrescu, Dr. Wolfgang Kathe, Adriana Morea, Maria Mihul, Michael Klemens, Dr. Florin Pacurar, Horatiu Popa, Razvan Popa, Bobby Pelger, Gârda Nicoleta, Dr. Susanne Schmitt, Luminita Tanasie Architects for Humanity: Chris Medland Babes-Bolyai University Cluj-Napoca (UBB) represented by Prof. Dr. Laszlo Rakosy Community Gârda de Sus, represented by the major Marin Virciu Darwin Initiative, London Drying (data collection): Bîte Daniela, Broscăţan Călin, Câmpean Sorin, Cosma Ramona, Dumitrescu Valentin, Feneşan Iulia, Gârda Nicoleta, Klemens Michael, Morea Adriana, Neag Cristina, Păcurar Adriana, Paşca Aniela, Pelger Bogdan, Rotar Bogdan, Spătăceanu Lucia, Tudose Sorina Ethnography: Dr. -

Chapter Vii Table of Contents
CHAPTER VII TABLE OF CONTENTS VII. APPENDICES AND REFERENCES CITED........................................................................1 Appendix 1: Description of Vegetation Databases......................................................................1 Appendix 2: Suggested Stocking Levels......................................................................................8 Appendix 3: Known Plants of the Desolation Watershed.........................................................15 Literature Cited............................................................................................................................25 CHAPTER VII - APPENDICES & REFERENCES - DESOLATION ECOSYSTEM ANALYSIS i VII. APPENDICES AND REFERENCES CITED Appendix 1: Description of Vegetation Databases Vegetation data for the Desolation ecosystem analysis was stored in three different databases. This document serves as a data dictionary for the existing vegetation, historical vegetation, and potential natural vegetation databases, as described below: • Interpretation of aerial photography acquired in 1995, 1996, and 1997 was used to characterize existing (current) conditions. The 1996 and 1997 photography was obtained after cessation of the Bull and Summit wildfires in order to characterize post-fire conditions. The database name is: 97veg. • Interpretation of late-1930s and early-1940s photography was used to characterize historical conditions. The database name is: 39veg. • The potential natural vegetation was determined for each polygon in the analysis -

THE USE of BIOTECHNOLOGY for SUPPLYING of PLANT MATERIAL for TRADITIONAL CULTURE of MEDICINAL, RARE SPECIES Arnica Montana L
Lucrări Ştiinţifice – vol. 57 (1) 2014, seria Agronomie THE USE OF BIOTECHNOLOGY FOR SUPPLYING OF PLANT MATERIAL FOR TRADITIONAL CULTURE OF MEDICINAL, RARE SPECIES Arnica montana L. Iuliana PANCIU1, Irina HOLOBIUC2, Rodica CĂTANĂ2 e-mail: [email protected] Abstract Taking into account the importance of Arnica montana, the attempts to improve the culture technologies are justified. Our study had the aim to optimize in vitro plant multiplication and growth as a source of plants for traditional culture in this species. Aseptic germinated seedlings were used as explants, apical meristem being the origin of the direct morphogenesis process. For induction of regeneration, to promote plant growth and rooting, we used some combination of growth factors and supplements as ascorbic acid, glutamine, PVP and active charcoal added in culture media based on MS formula. We improved the efficiency of micropropagation, the best values were recorded on variant supplemented with PVP –.7 regenerants/explant in the first 4 weeks and increasing at 17/ initial explant ( mean 14.62) after 8 weeks. Concerning the germination capacity of the seeds scored after 2 weeks in sterile condition, the rate was 47.76 and in non-sterile conditions, the rate varied depending of the substrate used. Comparing to the plants obtained through traditional seeds germination, in vitro plants grew faster and were more vigourously. The micropropagation protocol in Arnica montana L. allowed us to regenerate healthy, developed and rooted plants in the second subculture cycle. This in vitro methodology can provide plant material for initiation of a conventional culture after acclimatization of the obtained vitroplants. -
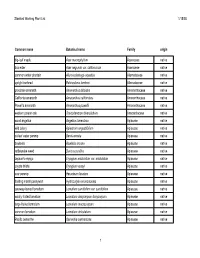
Plant List for Web Page
Stanford Working Plant List 1/15/08 Common name Botanical name Family origin big-leaf maple Acer macrophyllum Aceraceae native box elder Acer negundo var. californicum Aceraceae native common water plantain Alisma plantago-aquatica Alismataceae native upright burhead Echinodorus berteroi Alismataceae native prostrate amaranth Amaranthus blitoides Amaranthaceae native California amaranth Amaranthus californicus Amaranthaceae native Powell's amaranth Amaranthus powellii Amaranthaceae native western poison oak Toxicodendron diversilobum Anacardiaceae native wood angelica Angelica tomentosa Apiaceae native wild celery Apiastrum angustifolium Apiaceae native cutleaf water parsnip Berula erecta Apiaceae native bowlesia Bowlesia incana Apiaceae native rattlesnake weed Daucus pusillus Apiaceae native Jepson's eryngo Eryngium aristulatum var. aristulatum Apiaceae native coyote thistle Eryngium vaseyi Apiaceae native cow parsnip Heracleum lanatum Apiaceae native floating marsh pennywort Hydrocotyle ranunculoides Apiaceae native caraway-leaved lomatium Lomatium caruifolium var. caruifolium Apiaceae native woolly-fruited lomatium Lomatium dasycarpum dasycarpum Apiaceae native large-fruited lomatium Lomatium macrocarpum Apiaceae native common lomatium Lomatium utriculatum Apiaceae native Pacific oenanthe Oenanthe sarmentosa Apiaceae native 1 Stanford Working Plant List 1/15/08 wood sweet cicely Osmorhiza berteroi Apiaceae native mountain sweet cicely Osmorhiza chilensis Apiaceae native Gairdner's yampah (List 4) Perideridia gairdneri gairdneri Apiaceae -

National Wetlands Inventory Map Report for Quinault Indian Nation
National Wetlands Inventory Map Report for Quinault Indian Nation Project ID(s): R01Y19P01: Quinault Indian Nation, fiscal year 2019 Project area The project area (Figure 1) is restricted to the Quinault Indian Nation, bounded by Grays Harbor Co. Jefferson Co. and the Olympic National Park. Appendix A: USGS 7.5-minute Quadrangles: Queets, Salmon River West, Salmon River East, Matheny Ridge, Tunnel Island, O’Took Prairie, Thimble Mountain, Lake Quinault West, Lake Quinault East, Taholah, Shale Slough, Macafee Hill, Stevens Creek, Moclips, Carlisle. • < 0. Figure 1. QIN NWI+ 2019 project area (red outline). Source Imagery: Citation: For all quads listed above: See Appendix A Citation Information: Originator: USDA-FSA-APFO Aerial Photography Field Office Publication Date: 2017 Publication place: Salt Lake City, Utah Title: Digital Orthoimagery Series of Washington Geospatial_Data_Presentation_Form: raster digital data Other_Citation_Details: 1-meter and 1-foot, Natural Color and NIR-False Color Collateral Data: . USGS 1:24,000 topographic quadrangles . USGS – NHD – National Hydrography Dataset . USGS Topographic maps, 2013 . QIN LiDAR DEM (3 meter) and synthetic stream layer, 2015 . Previous National Wetlands Inventories for the project area . Soil Surveys, All Hydric Soils: Weyerhaeuser soil survey 1976, NRCS soil survey 2013 . QIN WET tables, field photos, and site descriptions, 2016 to 2019, Janice Martin, and Greg Eide Inventory Method: Wetland identification and interpretation was done “heads-up” using ArcMap versions 10.6.1. US Fish & Wildlife Service (USFWS) National Wetlands Inventory (NWI) mapping contractors in Portland, Oregon completed the original aerial photo interpretation and wetland mapping. Primary authors: Nicholas Jones of SWCA Environmental Consulting. 100% Quality Control (QC) during the NWI mapping was provided by Michael Holscher of SWCA Environmental Consulting. -

Vascular Plant Inventory of Mount Rainier National Park
National Park Service U.S. Department of the Interior Natural Resource Program Center Vascular Plant Inventory of Mount Rainier National Park Natural Resource Technical Report NPS/NCCN/NRTR—2010/347 ON THE COVER Mount Rainier and meadow courtesy of 2007 Mount Rainier National Park Vegetation Crew Vascular Plant Inventory of Mount Rainier National Park Natural Resource Technical Report NPS/NCCN/NRTR—2010/347 Regina M. Rochefort North Cascades National Park Service Complex 810 State Route 20 Sedro-Woolley, Washington 98284 June 2010 U.S. Department of the Interior National Park Service Natural Resource Program Center Fort Collins, Colorado The National Park Service, Natural Resource Program Center publishes a range of reports that address natural resource topics of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Technical Report Series is used to disseminate results of scientific studies in the physical, biological, and social sciences for both the advancement of science and the achievement of the National Park Service mission. The series provides contributors with a forum for displaying comprehensive data that are often deleted from journals because of page limitations. All manuscripts in the series receive the appropriate level of peer review to ensure that the information is scientifically credible, technically accurate, appropriately written for the intended audience, and designed and published in a professional manner. This report received informal peer review by subject-matter experts who were not directly involved in the collection, analysis, or reporting of the data. -

Download The
SYSTEMATICA OF ARNICA, SUBGENUS AUSTROMONTANA AND A NEW SUBGENUS, CALARNICA (ASTERACEAE:SENECIONEAE) by GERALD BANE STRALEY B.Sc, Virginia Polytechnic Institute, 1968 M.Sc, Ohio University, 1974 A THESIS SUBMITTED IN PARTIAL FULFILMENT OF THE REQUIREMENTS OF THE DEGREE OF DOCTOR OF PHILOSOPHY in THE FACULTY OF GRADUATE STUDIES (Department of Botany) We accept this thesis as conforming to the required standard THE UNIVERSITY OF BRITISH COLUMBIA March 1980 © Gerald Bane Straley, 1980 In presenting this thesis in partial fulfilment of the requirements for an advanced degree at the University of British Columbia, I agree that the Library shall make it freely available for reference and study. I further agree that permission for extensive copying of this thesis for scholarly purposes may be granted by the Head of my Department or by his representatives. It is understood that copying or publication of this thesis for financial gain shall not be allowed without my written permission. Department nf Botany The University of British Columbia 2075 Wesbrook Place Vancouver, Canada V6T 1W5 26 March 1980 ABSTRACT Seven species are recognized in Arnica subgenus Austromontana and two species in a new subgenus Calarnica based on a critical review and conserva• tive revision of the species. Chromosome numbers are given for 91 populations representing all species, including the first reports for Arnica nevadensis. Results of apomixis, vegetative reproduction, breeding studies, and artifi• cial hybridizations are given. Interrelationships of insect pollinators, leaf miners, achene feeders, and floret feeders are presented. Arnica cordifolia, the ancestral species consists largely of tetraploid populations, which are either autonomous or pseudogamous apomicts, and to a lesser degree diploid, triploid, pentaploid, and hexaploid populations. -

Chloroplast DNA Evidence for a North American Origin of the Hawaiian
Proc, Natl. Acad. Sci. USA Vol. 88, pp. 1840-1843, March 1991 Evolution Chloroplast DNA evidence for a North American origin of the Hawaiian silversword alliance (Asteraceae) (insular evolution/adaptive radiation/biogeography/long-distance dispersal/phylogenetics) BRUCE G. BALDWIN*t, DONALD W. KYHOS*, JAN DVORAKO, AND GERALD D. CARR§ Departments of *Botany and tAgronomy and Range Science, University of California, Davis, CA 95616; and §Department of Botany, University of Hawaii, Honolulu, HI 96822 Communicated by Peter H. Raven, November 30, 1990 (received for review August 1, 1990) ABSTRACT Chloroplast DNA restriction-site compari- sity, structural (4, 5), biosystematic (6), allozymic (7), and sons were made among 24 species of the Hawaiian silversword chloroplast DNA (cpDNA) (8) data indicate the silversword alliance (Argyroxiphium, Dubautia, and Wilkesia) and 7 species alliance originated from a single colonizing species. of North American perennial tarweeds in Adenothamnus, Ma- Carlquist (1) presented convincing anatomical evidence dia, Raillardella, and Railkrdiopsis (Asteraceae-Madiinae). indicating taxonomic alignment of the Hawaiian silversword These data and results from intergeneric hybridization indi- alliance with the almost exclusively herbaceous American cated surprisingly close genetic affinity of the monophyletic Madiinae or tarweeds. Gray (9) earlier suggested such affinity Hawaiian group to two diploid species of montane perennial for Argyroxiphium, which was disputed by Keck (10) based herbs in California, Madia bolanderi and Raillardiopsis muiri. on presumed morphological dissimilarities and the magnitude Of 117 restriction-site mutations shared among a subset of two of the oceanic barrier to migration. Herein, we compare or more accessions, more than one-fifth (25 mutations) sepa- cpDNA restriction sites between the silversword alliance and rated the silversword alliance, M.