Automated Taxonomic Identification of Insects with Expert-Level
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Monte L. Bean Life Science Museum Brigham Young University Provo, Utah 84602 PBRIA a Newsletter for Plecopterologists
No. 10 1990/1991 Monte L. Bean Life Science Museum Brigham Young University Provo, Utah 84602 PBRIA A Newsletter for Plecopterologists EDITORS: Richard W, Baumann Monte L. Bean Life Science Museum Brigham Young University Provo, Utah 84602 Peter Zwick Limnologische Flußstation Max-Planck-Institut für Limnologie, Postfach 260, D-6407, Schlitz, West Germany EDITORIAL ASSISTANT: Bonnie Snow REPORT 3rd N orth A merican Stonefly S ymposium Boris Kondratieff hosted an enthusiastic group of plecopterologists in Fort Collins, Colorado during May 17-19, 1991. More than 30 papers and posters were presented and much fruitful discussion occurred. An enjoyable field trip to the Colorado Rockies took place on Sunday, May 19th, and the weather was excellent. Boris was such a good host that it was difficult to leave, but many participants traveled to Santa Fe, New Mexico to attend the annual meetings of the North American Benthological Society. Bill Stark gave us a way to remember this meeting by producing a T-shirt with a unique “Spirit Fly” design. ANNOUNCEMENT 11th International Stonefly Symposium Stan Szczytko has planned and organized an excellent symposium that will be held at the Tree Haven Biological Station, University of Wisconsin in Tomahawk, Wisconsin, USA. The registration cost of $300 includes lodging, meals, field trip and a T- Shirt. This is a real bargain so hopefully many colleagues and friends will come and participate in the symposium August 17-20, 1992. Stan has promised good weather and good friends even though he will not guarantee that stonefly adults will be collected during the field trip. Printed August 1992 1 OBITUARIES RODNEY L. -

Download .PDF(1340
Stark, Bill P. and Stephen Green. 2011. Eggs of western Nearctic Acroneuriinae (Plecoptera: Perlidae). Illiesia, 7(17):157-166. Available online: http://www2.pms-lj.si/illiesia/Illiesia07-17.pdf EGGS OF WESTERN NEARCTIC ACRONEURIINAE (PLECOPTERA: PERLIDAE) Bill P. Stark1 and Stephen Green2 1,2 Box 4045, Department of Biology, Mississippi College, Clinton, Mississippi, U.S.A. 39058 1 E-mail: [email protected] 2 E-mail: [email protected] ABSTRACT Eggs for western Nearctic acroneuriine species of Calineuria Ricker, Doroneuria Needham & Claassen and Hesperoperla Banks are examined and redescribed based on scanning electron microscopy images taken from specimens collected from a substantial portion of each species range. Within genera, species differences in egg morphology are small and not always useful for species recognition, however eggs from one population of Calineuria are significantly different from those found in other populations and this population is given informal recognition as a possible new species. Keywords: Plecoptera, Calineuria, Doroneuria, Hesperoperla, Egg morphology, Western Nearctic INTRODUCTION occur in the region (Baumann & Olson 1984; Scanning electron microscopy (SEM) is often used Kondratieff & Baumann 2002; Stark 1989; Stark & to elucidate chorionic features for stoneflies (e.g. Gaufin 1976; Stark & Kondratieff 2004; Zuellig et al. Baumann 1973; Grubbs 2005; Isobe 1988; Kondratieff 2006). SEM images for eggs of the primary western 2004; Kondratieff & Kirchner 1996; Nelson 2000; acroneuriine genera, Calineuria Ricker, Doroneuria Sivec & Stark 2002; 2008; Stark & Nelson 1994; Stark Needham & Claassen and Hesperoperla Banks include & Szczytko 1982; 1988; Szczytko & Stewart 1979) and single images for each of these genera in Stark & Nearctic Perlidae were among the earliest stoneflies Gaufin (1976), three images of Hesperoperla hoguei to be studied with this technique (Stark & Gaufin Baumann & Stark (1980) and three images of H. -

Circularly Polarized Reflection from the Scarab Beetle Chalcothea Smaragdina: Rsfs.Royalsocietypublishing.Org Light Scattering by a Dual Photonic Structure
Circularly polarized reflection from the scarab beetle Chalcothea smaragdina: rsfs.royalsocietypublishing.org light scattering by a dual photonic structure Luke T. McDonald1,2, Ewan D. Finlayson1, Bodo D. Wilts3 and Pete Vukusic1 Research 1Department of Physics and Astronomy, University of Exeter, Stocker Road, Exeter EX4 4QL, UK 2School of Biological, Earth and Environmental Sciences, University College Cork, North Mall Campus, Cork, Cite this article: McDonald LT, Finlayson ED, Republic of Ireland Wilts BD, Vukusic P. 2017 Circularly polarized 3Adolphe Merkle Institute, University of Fribourg, Chemin des Verdiers 4, 1700 Fribourg, Switzerland reflection from the scarab beetle Chalcothea LTM, 0000-0003-0896-1415; EDF, 0000-0002-0433-5313; BDW, 0000-0002-2727-7128 smaragdina: light scattering by a dual photonic structure. Interface Focus 7: 20160129. Helicoidal architectures comprising various polysaccharides, such as chitin http://dx.doi.org/10.1098/rsfs.2016.0129 and cellulose, have been reported in biological systems. In some cases, these architectures exhibit stunning optical properties analogous to ordered cholesteric liquid crystal phases. In this work, we characterize the circularly One contribution of 17 to a theme issue polarized reflectance and optical scattering from the cuticle of the beetle ‘Growth and function of complex forms in Chalcothea smaragdina (Coleoptera: Scarabaeidae: Cetoniinae) using optical biological tissue and synthetic self-assembly’. experiments, simulations and structural analysis. The selective reflection of left-handed circularly polarized light is attributed to a Bouligand-type Subject Areas: helicoidal morphology within the beetle’s exocuticle. Using electron microscopy to inform electromagnetic simulations of this anisotropic strati- biomaterials fied medium, the inextricable connection between the colour appearance of C. -

Annual Newsletter and Bibliography of the International Society of Plecopterologists PERLA NO. 28, 2010
PERLA Annual Newsletter and Bibliography of The International Society of Plecopterologists Pteronarcella regularis (Hagen), Mt. Shasta City Park, California, USA. Photograph by Bill P. Stark PERLA NO. 28, 2010 Department of Bioagricultural Sciences and Pest Management Colorado State University Fort Collins, Colorado 80523 USA PERLA Annual Newsletter and Bibliography of the International Society of Plecopterologists Available on Request to the Managing Editor MANAGING EDITOR: Boris C. Kondratieff Department of Bioagricultural Sciences And Pest Management Colorado State University Fort Collins, Colorado 80523 USA E-mail: [email protected] EDITORIAL BOARD: Richard W. Baumann Department of Biology and Monte L. Bean Life Science Museum Brigham Young University Provo, Utah 84602 USA E-mail: [email protected] J. Manuel Tierno de Figueroa Dpto. de Biología Animal Facultad de Ciencias Universidad de Granada 18071 Granada, SPAIN E-mail: [email protected] Kenneth W. Stewart Department of Biological Sciences University of North Texas Denton, Texas 76203, USA E-mail: [email protected] Shigekazu Uchida Aichi Institute of Technology 1247 Yagusa Toyota 470-0392, JAPAN E-mail: [email protected] Peter Zwick Schwarzer Stock 9 D-36110 Schlitz, GERMANY E-mail: [email protected] 2 TABLE OF CONTENTS Subscription policy……………………………………………………………………….4 Publication of the Proceedings of the International Joint Meeting on Ephemeroptera and Plecoptera 2008…………………………………….………………….………….…5 Ninth North American Plecoptera Symposium………………………………………….6 -

Coleoptera: Cetoniidae) and Their Damages on Peach Fruits in Orchards of Northern Dalmatia, Croatia
CORE Metadata, citation and similar papers at core.ac.uk Entomol. Croat. 2009, Vol. 13. Num. 2: 7-20 ISSN 1330-6200 IzvORNI zNANSTvENI čLANCI ORIGINAL SCIENTIfIC PAPER Fauna Of THE Cetoniid BEETLES (Coleoptera: Cetoniidae) AND THEIR DAMAGES ON PEACH fRUITS IN ORCHARDS Of NorthERN Dalmatia, Croatia Josip RAžOV¹, Božena BARIĆ² & Moreno DUTTO³ ¹ University of Zadar, Department of Mediterranean Agriculture and Aquaculture, Mihovila Pavlinovica bb, 23000 Zadar, Croatia; e-mail: [email protected] ² Faculty of Agronomy, University of Zagreb, Svetošimunska 25, 10000 Zagreb, Croatia; e-mail: [email protected] ³ Sezione Entomologia Museo Civico Storia Naturale, Carmagnola, Italy; e-mail: [email protected] Accepted: June 29th 2009 The beetles Cetonia aurata and Potosia cuprea belonging to the subfamily Cetoniinae (Coleoptera: Cetoniidae) are present in peach orchards in Northern Dalmatia, Ravni kotari region. They are often described as flower pest (“Rose chafers, flower beetles”), and are thought not to be significant as fruit pests. However, during the last ten years some serious damage to fruit has been observed. Since this damage occurs when the fruits are ripening, insecticides cannot be used. There are no literature data about the amount of the damage or how to monitor the damage. This paper describes our monitoring of the population dynamics of the Cetonia aurata and Potosia cuprea, and the method for calculating the damage to fruit suitable for the orchards in this area. The study was conducted during the spring and summer of the year 2005, 2006 and 2007 in the Ravni kotari region, near the villages of Prkos and Smilčić. We used Csalomon® VARb3k funnel traps. -

The Diversity of the Family Cetoniidae (Coleoptera: Scarabaeoidea) of Mountain Ozren (Bosnia and Herzegovina)
Acta entomologica serbica, 2013, 18(1/2): 55-67 UDC 595.764(497.6) THE DIVERSITY OF THE FAMILY CETONIIDAE (COLEOPTERA: SCARABAEOIDEA) OF MOUNTAIN OZREN (BOSNIA AND HERZEGOVINA) MIRZETA KAŠIĆ-LELO*, SUVAD LELO and ADI VESNIĆ Faculty of Natural Sciences and Mathematics, Biology Department Zmaja od Bosne 33 71000 Sarajevo, Bosnia and Herzegovina * E-mail: [email protected] Abstract From April to September of 2008 and 2009 samples of family Cetoniidae Leach, 1815 were collected on the vertical profile of the southern slopes of Mount Ozren near Sarajevo. Research was conducted at six selected localities: Orlovac, Nahorevo, Čavljak, Skakvac, Bukovik and Crepoljsko. After 41 field investigations, 594 individuals were collected and determined as representatives of seven species. Collected data were processed with statistical software Biodiversity Pro and with options Diversity/Compare diversities: Alpha index; Berger-Parker index; Simpsons index; Margaleff index; Mackintosh index; Bray-Curtis Cluster Analysis: Single Link. By comparison of data it was concluded that Skakavac represents the typical locality for southern slopes of Mount Ozren. The species Oxythyrea funesta (Poda, 1761) is the most numerous faunistic element that was found. KEY WORDS: Cetoniidae, fauna, biodiversity, Ozren, Sarajevo, Bosnia and Herzegovina Introduction After World War II the most important data on flower chafer Cetoniidae Leach, 1815 of Bosnia and Herzegovina were given by Rene MIKŠIĆ who investigated the area of Bosnia and Herzegovina, the entire Balkan Peninsula and most of the Palaearctic (MIKŠIĆ, 1950, 1953, 1956, 1958, 1962, 1965, 1970, 1976, 1977, 1980, 1982, 1987, e.g.). What little research done on this group after the war events in Bosnia and Herzegovina from 1992-1995 was undertaken mostly by authors of this paper (LELO, 2000, 2003; LELO & KAŠIĆ-LELO, 2006a, 2006b, 2007; LELO & ŠKRIJELJ, 2001; KAŠIĆ-LELO, 2005, 2011; KAŠIĆ-LELO & LELO, 2005, 2006, 2007, 2009a, 2009b; KAŠIĆ-LELO et al., 2006), but it has been so little that it can be said that no 56 M. -
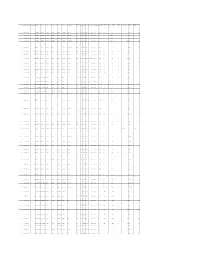
Appendix D: Summary of Accepted ECOTOX Papers
Effect Dur Dur Unit Dur Dur Unit Conc Conc Units Conc Value1 Purity CAS Number Chemical Name Species Number Phylum Class Order Family Genus Species Common Name Group Effect Meas Endpt1 Endpt2 Habitat Plant/Animal Media Orig Orig Preferred Preferred Type Conc Value1 Orig Orig Adjusted 1 298044 Disulfoton 4510 Chordata Mammalia Rodentia Muridae Rattus norvegicus Norway rat BEH AVO STIM NOAEL terrestrial Animal NONE 30 d 30 d F 2 mg/kg/d 1.96 2 298044 Disulfoton 351 Chordata Actinopterygii Perciformes Anabantidae Anabas testudineus Climbing perch BCM BCM LIPD LOAEL aquatic Animal FW 1 h 4.17E-02 d A 4 mg/L 4 3 298044 Disulfoton 351 Chordata Actinopterygii Perciformes Anabantidae Anabas testudineus Climbing perch BCM BCM PRCO LOAEL aquatic Animal FW 1 h 4.17E-02 d A 4 mg/L 4 4 298044 Disulfoton 351 Chordata Actinopterygii Perciformes Anabantidae Anabas testudineus Climbing perch BCM BCM LIPD LOAEL aquatic Animal FW 1 h 4.17E-02 d A 10.5 mg/L 10.5 5 298044 Disulfoton 4913 Chordata Mammalia Rodentia Muridae Mus musculus House mouse BCM BCM HXBT LOAEL terrestrial Animal NONE 3 d 3 d F 35.1 uM/kg 35.1 6 298044 Disulfoton 4510 Chordata Mammalia Rodentia Muridae Rattus norvegicus Norway rat BCM BCM GBCM LOAEL terrestrial Animal NONE 24 h 1 d F 0.26 mg/kg 0.234 7 298044 Disulfoton 4510 Chordata Mammalia Rodentia Muridae Rattus norvegicus Norway rat BCM BCM GBCM IC50 terrestrial Animal NONE 1 wk 7 d F 0.6 ppm 0.6 8 298044 Disulfoton 4510 Chordata Mammalia Rodentia Muridae Rattus norvegicus Norway rat BCM BCM GBCM NOAEL LOAEL terrestrial Animal NONE -

<I>Trichiotinus Rufobrunneus</I>
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Center for Systematic Entomology, Gainesville, Insecta Mundi Florida 2018 A remarkable teratological specimen of Trichiotinus rufobrunneus (Casey) (Coleoptera: Scarabaeidae: Cetoniinae: Trichiini) Héctor Jaime Gasca-Álvarez Corporacion Sentido Natural, [email protected] Paul E. Skelley Florida Department of Agriculture and Consumer Services, [email protected] Cuauhtemoc Deloya Instituto de Ecología, A.C., [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/insectamundi Part of the Ecology and Evolutionary Biology Commons, and the Entomology Commons Gasca-Álvarez, Héctor Jaime; Skelley, Paul E.; and Deloya, Cuauhtemoc, "A remarkable teratological specimen of Trichiotinus rufobrunneus (Casey) (Coleoptera: Scarabaeidae: Cetoniinae: Trichiini)" (2018). Insecta Mundi. 1175. http://digitalcommons.unl.edu/insectamundi/1175 This Article is brought to you for free and open access by the Center for Systematic Entomology, Gainesville, Florida at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Insecta Mundi by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. December 28 2018 INSECTA 0680 1–5 urn:lsid:zoobank.org:pub:6849188C-81E7-45E8-86C4-AB- A Journal of World Insect Systematics CB481A61EE MUNDI 0680 A remarkable teratological specimen of Trichiotinus rufobrunneus (Casey) (Coleoptera: Scarabaeidae: Cetoniinae: Trichiini) Héctor Jaime -

A Trait-Based Approach Laura Roquer Beni Phd Thesis 2020
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Pollinator communities and pollination services in apple orchards: a trait-based approach Laura Roquer Beni PhD Thesis 2020 Pollinator communities and pollination services in apple orchards: a trait-based approach Tesi doctoral Laura Roquer Beni per optar al grau de doctora Directors: Dr. Jordi Bosch i Dr. Anselm Rodrigo Programa de Doctorat en Ecologia Terrestre Centre de Recerca Ecològica i Aplicacions Forestals (CREAF) Universitat de Autònoma de Barcelona Juliol 2020 Il·lustració de la portada: Gala Pont @gala_pont Al meu pare, a la meva mare, a la meva germana i al meu germà Acknowledgements Se’m fa impossible resumir tot el que han significat per mi aquests anys de doctorat. Les qui em coneixeu més sabeu que han sigut anys de transformació, de reptes, d’aprendre a prioritzar sense deixar de cuidar allò que és important. Han sigut anys d’equilibris no sempre fàcils però molt gratificants. Heu sigut moltes les persones que m’heu acompanyat, d’una manera o altra, en el transcurs d’aquest projecte de creixement vital i acadèmic, i totes i cadascuna de vosaltres, formeu part del resultat final. -

INSECTA MUNDI a Journal of World Insect Systematics
INSECTA MUNDI A Journal of World Insect Systematics 0061 Synopsis of the Oryctini (Coleoptera: Scarabaeidae: Dynastinae) from the Brazilian Amazon Héctor Jaime Gasca Alvarez Instituto Nacional de Pesquisas da Amazônia-INPA Coordenação de Pesquisas em Entomologia Av. André Araújo, 2936-Petrópolis CEP 69011-970 Manaus, Amazonas, Brazil Claudio Ruy Vasconcelos da Fonseca Instituto Nacional de Pesquisas da Amazônia-INPA Coordenação de Pesquisas em Entomologia Av. André Araújo, 2936-Petrópolis CEP 69011-970 Manaus, Amazonas, Brazil Brett C. Ratcliffe University of Nebraska State Museum W436 Nebraska Hall Lincoln, NE 68588-0514, USA Date of Issue: December 5, 2008 CENTER FOR SYSTEMATIC ENTOMOLOGY, INC., Gainesville, FL Héctor Jaime Gasca Alvarez, Claudio Ruy Vasconcelos da Fonseca, and Brett C. Ratcliffe Synopsis of the Oryctini (Coleoptera: Scarabaeidae: Dynastinae) from the Brazilian Amazon Insecta Mundi 0061: 1-62 Published in 2008 by Center for Systematic Entomology, Inc. P. O. Box 141874 Gainesville, FL 32614-1874 U. S. A. http://www.centerforsystematicentomology.org/ Insecta Mundi is a journal primarily devoted to insect systematics, but articles can be published on any non-marine arthropod taxon. Manuscripts considered for publication include, but are not limited to, systematic or taxonomic studies, revisions, nomenclatural changes, faunal studies, book reviews, phylo- genetic analyses, biological or behavioral studies, etc. Insecta Mundi is widely distributed, and refer- enced or abstracted by several sources including the Zoological Record, CAB Abstracts, etc. As of 2007, Insecta Mundi is published irregularly throughout the year, not as quarterly issues. As manuscripts are completed they are published and given an individual number. Manuscripts must be peer reviewed prior to submission, after which they are again reviewed by the editorial board to insure quality. -

In Baltic Amber 85
Overview and descriptions of fossil stoneflies (Plecoptera) in Baltic amber 85 Entomologie heute 22 (2010): 85-97 Overview and Descriptions of Fossil Stoneflies (Plecoptera) in Baltic Amber Übersicht und Beschreibungen von fossilen Steinfliegen (Plecoptera) im Baltischen Bernstein CELESTINE CARUSO & WILFRIED WICHARD Summary: Three new fossil species of stoneflies (Plecoptera: Nemouridae and Leuctridae) from Eocene Baltic amber are being described: Zealeuctra cornuta n. sp., Lednia zilli n. sp., and Podmosta attenuata n. sp.. Extant species of these three genera are found in Eastern Asia and in the Nearctic region. It is very probably that the genera must have been widely spread across the northern hemisphere in the Cretaceous period, before Europe was an archipelago in Eocene. The current state of knowledge about the seventeen Plecoptera species of Baltic amber is shortly presented. Due to discovered homonymies, the following nomenclatural corrections are proposed: Leuctra fusca Pictet, 1856 in Leuctra electrofusca Caruso & Wichard, 2010 and Nemoura affinis Berendt, 1856 in Nemoura electroaffinis Caruso & Wichard, 2010. Keywords: Fossil insects, fossil Plecoptera, Eocene, paleobiogeography Zusammenfassung: In dieser Arbeit werden drei neue fossile Steinfliegen-Arten (Plecoptera: Nemouridae und Leuctridae) des Baltischen Bernsteins beschrieben: Zealeuctra cornuta n. sp., Lednia zilli n. sp., Podmosta attenuata n. sp.. Rezente Arten der drei Gattungen sind in Ostasien und in der nearktischen Region nachgewiesen. Sehr wahrscheinlich breiteten sich die Gattungen in der Krei- dezeit über die nördliche Hemisphäre aus, noch bevor Europa im Eozän ein Archipel war. Der gegenwärtige Kenntnisstand über die siebzehn Plecoptera Arten des Baltischen Bernsteins wird kurz dargelegt. Wegen bestehender Homonymien werden folgende nomenklatorische Korrekturen vorgenommen: Leuctra fusca Pictet, 1856 in Leuctra electrofusca Caruso & Wichard, 2010 und Nemoura affinis Berendt, 1856 in Nemoura electroaffinis Caruso & Wichard, 2010.