A Molecular Phylogeny for Marine Turtles: Trait Mapping, Rate
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
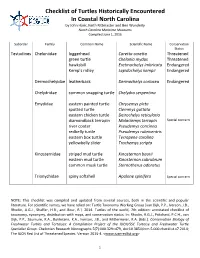
N.C. Turtles Checklist
Checklist of Turtles Historically Encountered In Coastal North Carolina by John Hairr, Keith Rittmaster and Ben Wunderly North Carolina Maritime Museums Compiled June 1, 2016 Suborder Family Common Name Scientific Name Conservation Status Testudines Cheloniidae loggerhead Caretta caretta Threatened green turtle Chelonia mydas Threatened hawksbill Eretmochelys imbricata Endangered Kemp’s ridley Lepidochelys kempii Endangered Dermochelyidae leatherback Dermochelys coriacea Endangered Chelydridae common snapping turtle Chelydra serpentina Emydidae eastern painted turtle Chrysemys picta spotted turtle Clemmys guttata eastern chicken turtle Deirochelys reticularia diamondback terrapin Malaclemys terrapin Special concern river cooter Pseudemys concinna redbelly turtle Pseudemys rubriventris eastern box turtle Terrapene carolina yellowbelly slider Trachemys scripta Kinosternidae striped mud turtle Kinosternon baurii eastern mud turtle Kinosternon subrubrum common musk turtle Sternotherus odoratus Trionychidae spiny softshell Apalone spinifera Special concern NOTE: This checklist was compiled and updated from several sources, both in the scientific and popular literature. For scientific names, we have relied on: Turtle Taxonomy Working Group [van Dijk, P.P., Iverson, J.B., Rhodin, A.G.J., Shaffer, H.B., and Bour, R.]. 2014. Turtles of the world, 7th edition: annotated checklist of taxonomy, synonymy, distribution with maps, and conservation status. In: Rhodin, A.G.J., Pritchard, P.C.H., van Dijk, P.P., Saumure, R.A., Buhlmann, K.A., Iverson, J.B., and Mittermeier, R.A. (Eds.). Conservation Biology of Freshwater Turtles and Tortoises: A Compilation Project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group. Chelonian Research Monographs 5(7):000.329–479, doi:10.3854/crm.5.000.checklist.v7.2014; The IUCN Red List of Threatened Species. -

Gopher Tortoise Demographic Responses to a Novel Disturbance Regime
The Journal of Wildlife Management 1–10; 2019; DOI: 10.1002/jwmg.21774 Research Article Gopher Tortoise Demographic Responses to a Novel Disturbance Regime HUNTER J. HOWELL ,1,2 Department of Biology, University of Miami, 1301 Memorial Drive, Coral Gables, FL 33146, USA BETSIE B. ROTHERMEL,2 Archbold Biological Station, 123 Main Drive, Venus, FL 33960, USA K. NICOLE WHITE, Daniel B. Warnell School of Forestry and Natural Resources, University of Georgia, 180 E Green Street, Athens, GA 30602, USA CHRISTOPHER A. SEARCY, Department of Biology, University of Miami, 1301 Memorial Drive, Coral Gables, FL 33146, USA ABSTRACT The long‐term viability of gopher tortoise (Gopherus polyphemus) populations is jeopardized by increased urbanization and habitat degradation owing to fire suppression. Because the species’ remaining natural habitats in the southeastern United States exist within a mosaic of anthropogenic land uses, it is important to understand demographic responses to contrasting land uses and habitat management regimes. We examined differences in demographic parameters among fire‐suppressed sandhill, restored sandhill, and former sandhill (i.e., ruderal) land use‐land cover (LULC) types at Archbold Biological Station in south‐ central Florida, USA. Using Program MARK, we estimated population size, and sex‐specific and LULC‐specific survivorship based on 6 years of mark‐recapture data. We also analyzed individual growth trajectories and clutch sizes to determine whether growth rates or reproductive output differed among LULC types. Tortoises in an open, ruderal field occurred at a higher density (7.79/ha) than in adjacent restored (1.43/ha) or fire‐suppressed (0.40/ha) sandhill. Despite this higher density, both adult survivorship and body size were significantly higher in the ruderal field. -

The Conservation Biology of Tortoises
The Conservation Biology of Tortoises Edited by Ian R. Swingland and Michael W. Klemens IUCN/SSC Tortoise and Freshwater Turtle Specialist Group and The Durrell Institute of Conservation and Ecology Occasional Papers of the IUCN Species Survival Commission (SSC) No. 5 IUCN—The World Conservation Union IUCN Species Survival Commission Role of the SSC 3. To cooperate with the World Conservation Monitoring Centre (WCMC) The Species Survival Commission (SSC) is IUCN's primary source of the in developing and evaluating a data base on the status of and trade in wild scientific and technical information required for the maintenance of biological flora and fauna, and to provide policy guidance to WCMC. diversity through the conservation of endangered and vulnerable species of 4. To provide advice, information, and expertise to the Secretariat of the fauna and flora, whilst recommending and promoting measures for their con- Convention on International Trade in Endangered Species of Wild Fauna servation, and for the management of other species of conservation concern. and Flora (CITES) and other international agreements affecting conser- Its objective is to mobilize action to prevent the extinction of species, sub- vation of species or biological diversity. species, and discrete populations of fauna and flora, thereby not only maintain- 5. To carry out specific tasks on behalf of the Union, including: ing biological diversity but improving the status of endangered and vulnerable species. • coordination of a programme of activities for the conservation of biological diversity within the framework of the IUCN Conserva- tion Programme. Objectives of the SSC • promotion of the maintenance of biological diversity by monitor- 1. -

Relative Abundance, Population Structure, and Habitat
University of Missouri, St. Louis IRL @ UMSL Theses Graduate Works 11-18-2010 Relative Abundance, Population Structure, and Habitat Utilization of the Alligator Snapping Turtle (Macrochelys temminckii) and Eastern Snapping Turtle (Chelydra serpentina) in southeastern Missouri Timothy Charles Lescher University of Missouri-St. Louis, [email protected] Follow this and additional works at: http://irl.umsl.edu/thesis Recommended Citation Lescher, Timothy Charles, "Relative Abundance, Population Structure, and Habitat Utilization of the Alligator Snapping Turtle (Macrochelys temminckii) and Eastern Snapping Turtle (Chelydra serpentina) in southeastern Missouri" (2010). Theses. 44. http://irl.umsl.edu/thesis/44 This Thesis is brought to you for free and open access by the Graduate Works at IRL @ UMSL. It has been accepted for inclusion in Theses by an authorized administrator of IRL @ UMSL. For more information, please contact [email protected]. Relative Abundance, Population Structure, and Habitat Utilization of the Alligator Snapping Turtle (Macrochelys temminckii) and Eastern Snapping Turtle (Chelydra serpentina) in southeastern Missouri Timothy C. Lescher B.S., Biology, University of Missouri-Columbia, 2001 A Thesis Submitted to the Graduate School at the University of Missouri-Saint Louis in partial fulfillment of the requirements for the degree Master’s of Science in Biology November 2010 Advisory Committee Zuleyma Tang-Martinez, Ph.D. Chair Jeffrey T. Briggler, Ph.D. Bette A. Loiselle, Ph.D. Copyright, Timothy C. Lescher, 2010 1 TABLE -

The Common Snapping Turtle, Chelydra Serpentina
The Common Snapping Turtle, Chelydra serpentina Rylen Nakama FISH 423: Olden 12/5/14 Figure 1. The Common Snapping Turtle, one of the most widespread reptiles in North America. Photo taken in Quebec, Canada. Image from https://www.flickr.com/photos/yorthopia/7626614760/. Classification Order: Testudines Family: Chelydridae Genus: Chelydra Species: serpentina (Linnaeus, 1758) Previous research on Chelydra serpentina (Phillips et al., 1996) acknowledged four subspecies, C. s. serpentina (Northern U.S. and Figure 2. Side profile of Chelydra serpentina. Note Canada), C. s. osceola (Southeastern U.S.), C. s. the serrated posterior end of the carapace and the rossignonii (Central America), and C. s. tail’s raised central ridge. Photo from http://pelotes.jea.com/AnimalFact/Reptile/snapturt.ht acutirostris (South America). Recent IUCN m. reclassification of chelonians based on genetic analyses (Rhodin et al., 2010) elevated C. s. rossignonii and C. s. acutirostris to species level and established C. s. osceola as a synonym for C. s. serpentina, thus eliminating subspecies within C. serpentina. Antiquated distinctions between the two formerly recognized North American subspecies were based on negligible morphometric variations between the two populations. Interbreeding in the overlapping range of the two populations was well documented, further discrediting the validity of the subspecies distinction (Feuer, 1971; Aresco and Gunzburger, 2007). Therefore, any emphasis of subspecies differentiation in the ensuing literature should be disregarded. Figure 3. Front-view of a captured Chelydra Continued usage of invalid subspecies names is serpentina. Different skin textures and the distinctive pink mouth are visible from this angle. Photo from still prevalent in the exotic pet trade for C. -

UC Davis UC Davis Previously Published Works
UC Davis UC Davis Previously Published Works Title Survival, Demography, and Growth of Gopher Tortoises (Gopherus polyphemus) from Three Study Sites with Different Management Histories Permalink https://escholarship.org/uc/item/9sc747h0 Journal JOURNAL OF WILDLIFE MANAGEMENT, 78(7) ISSN 0022-541X Authors Tuberville, Tracey D Todd, Brian D Hermann, Sharon M et al. Publication Date 2014-09-01 DOI 10.1002/jwmg.773 Peer reviewed eScholarship.org Powered by the California Digital Library University of California The Journal of Wildlife Management 78(7):1151–1160; 2014; DOI: 10.1002/jwmg.773 Research Article Survival, Demography, and Growth of Gopher Tortoises (Gopherus polyphemus) from Three Study Sites with Different Management Histories TRACEY D. TUBERVILLE,1 University of Georgia’s Savannah River Ecology Lab, Drawer E, Aiken, SC 29802, USA BRIAN D. TODD, Department of Wildlife, Fish, and Conservation Biology, University of California, Davis, One Shields Ave, Davis, CA 95616, USA SHARON M. HERMANN, Department of Biological Sciences, Auburn University, Auburn, AL 36849, USA WILLIAM K. MICHENER, University Libraries, 1312 Basehart SE, Albuquerque, NM 87106, USA CRAIG GUYER, Department of Biological Sciences, Auburn University, Auburn, AL 36849, USA ABSTRACT Recovery or sustainable management of wildlife populations often entails management of habitat on which they depend. In this regard, turtles pose unique conservation challenges because of their life histories. The combination of late maturity, low survival when young, and dependence on high adult survival suggests they may be slow to respond demographically to conventional habitat management. Thus, long- term studies are necessary to understand population dynamics and recovery potential in these species. -

Snapping Turtle, Chelydra Serpentina, Overland Movements Near the Southeastern Extent of Its Range David A
Georgia Journal of Science Volume 68 No. 2 Scholarly Contributions from the Article 11 Membership and Others 2010 Snapping Turtle, Chelydra serpentina, Overland Movements Near the Southeastern Extent of its Range David A. Steen [email protected] Sean C. Sterrett Aubrey M. Heupel Lora L. Smith Follow this and additional works at: https://digitalcommons.gaacademy.org/gjs Part of the Life Sciences Commons Recommended Citation Steen, David A.; Sterrett, Sean C.; Heupel, Aubrey M.; and Smith, Lora L. (2010) "Snapping Turtle, Chelydra serpentina, Overland Movements Near the Southeastern Extent of its Range," Georgia Journal of Science, Vol. 68, No. 2, Article 11. Available at: https://digitalcommons.gaacademy.org/gjs/vol68/iss2/11 This Research Articles is brought to you for free and open access by Digital Commons @ the Georgia Academy of Science. It has been accepted for inclusion in Georgia Journal of Science by an authorized editor of Digital Commons @ the Georgia Academy of Science. 196 Steen et al.: Snapping Turtle Overland Movements SNAPPING Turtle, CHELYDRA SERPENTINA, OVERLAND MOVEMENTS NEAR THE SOUTHEASTERN EXTENT OF ITS RANGE David A. Steen1,2*, Sean C. Sterrett2, Aubrey M. Heupel2 and Lora L. Smith2 1Auburn University, 331 Funchess Hall, Auburn, Alabama, 36849 2Joseph W. Jones Ecological Research Center Route 2, Box 2324, Newton, GA 39870 Institution at which work was completed: Joseph W. Jones Ecological Research Center * Corresponding author: [email protected] ABSTRACT Terrestrial movements of turtles are of interest due to the conserva- tion implications for this imperiled group and the general lack of information on this topic, particularly in wide-ranging species. -

North Louisiana Refuges Complex: Freshwater Turtle Inventory
NORTH LOUISIANA REFUGES COMPLEX: FRESHWATER TURTLE INVENTORY USFWS Award No: F17PX01556 John L. Carr, Aaron C. Johnson & J. Benjamin Grizzle November 2020 ACKNOWLEDGMENTS We thank the U.S. Fish and Wildlife Service Region 4 Inventory and Monitoring Branch for USFWS Award No. F17PX01556, “Freshwater Turtle Inventory of the North Louisiana Refuges Complex”. In addition, this report incorporates data from other, complementary projects that were funded by a variety of sources. This has been done in order to provide a more fulsome picture of knowledge on the turtle fauna of the refuges within the Complex. These other projects were funded in part by the Louisiana Department of Wildlife and Fisheries and the U.S. Fish and Wildlife Service, Division of Federal Aid, through the State Wildlife Grants Program (series of projects targeting the Alligator Snapping Turtle and map turtles). Other sources include data collected for grant activities funded by USGS-BRD (Cooperative Agreement No. 99CRAG0017), funding from the Louisiana Wildlife and Fisheries Foundation beginning in 1998, funding directly from LDWF (2000-2002), The Nature Conservancy (contract #LAFO_022309), Friends of Black Bayou, the Turtle Research Fund of the University of Louisiana at Monroe Foundation, and the Kitty DeGree Professorship in Biology (2011-2017). U.S. Fish and Wildlife personnel helped facilitate our work by granting access to all parts of the refuges at various times, and providing a series of Special Use Permits over 20+ years. We acknowledge Lee Fulton, Joe McGowan, Brett Hortman, Kelby Ouchley, and George Chandler for facilitating our work on refuges over the years; in particular, we thank Gypsy Hanks for long-sustained support and cooperation, especially during the course of the current project. -

Chelydra Serpentina, Chelydridae
WWW.IRCF.ORG/REPTILESANDAMPHIBIANSJOURNALTABLE OF CONTENTS IRCF REPTILES & AMPHIBIANSIRCF REPTILES • VOL15, N O& 4 AMPHIBIANS • DEC 2008 189 • 20(1):1–6 • MAR 2013 IRCF REPTILES & AMPHIBIANS CONSERVATION AND NATURAL HISTORY TABLE OF CONTENTS FEATURE ARTICLES Necrotized. Chasing Bullsnakes (Pituophis Facial catenifer sayi) in Wisconsin: Deformities in an Urban On the Road to Understanding the Ecology and Conservation of the Midwest’s Giant Serpent ...................... Joshua M. Kapfer 190 . The Shared History of Treeboas (Corallus grenadensis) and Humans on Grenada: PopulationA Hypothetical Excursion ............................................................................................................................of Common SnappingRobert W. Henderson Turtles 198 RESEARCH(Chelydra ARTICLES serpentina, Chelydridae) . The Texas Horned Lizard in Central and Western Texas ....................... Emily Henry, Jason Brewer, Krista Mougey, and Gad Perry 204 . The Knight Anole (Anolis equestris) in Florida .............................................fromBrian J. Camposano,Bronx, Kenneth L. Krysko, KevinNew M. Enge, Ellen M.York Donlan, and Michael Granatosky 212 ErikCONSERVATION Zeidler, New York ALERTWild! (www.ny-wild.com), Bronx, New York 10461 ([email protected]) . World’s Mammals in Crisis ............................................................................................................................................................. 220 . More Than Mammals ......................................................................................................................................................................Photographs -

The Alligator Snapping Turtle (Macrochelys Temminckii) Is the Largest Freshwater Turtle in North America
TheThe AlligatorAlligator SnappingSnapping TurtleTurtle [[MacrochelysMacrochelys ((MacroclemysMacroclemys)) temminckiitemminckii]:]: A review of ecology, life history, and conservation, with demographic analyses of the sustainability of take from wild populations a report to: Division of Scientific Authority United States Fish and Wildlife Service prepared by: Robert N. Reed, PhD Justin Congdon, PhD J. Whitfield Gibbons, PhD Savannah River Ecology Laboratory Drawer E Aiken SC 29802 USA ...Table of Contents... Executive summary ................................................................................................................................ 1 Introduction........................................................................................................................................... 2 Taxonomy, distribution, and systematics................................................................................................. 2 Ecology and Natural History ................................................................................................................... 3 Size and growth rates ............................................................................................................... 3 Longevity and age at first reproduction ..................................................................................... 3 Reproductive biology ................................................................................................................ 4 Survivorship ............................................................................................................................ -

Nesting Ecology and Habitat Use of Chelydra Serpentina in an Area Modified by Agricultural and Industrial Activity
Herpetological Conservation and Biology 12:292–306. Submitted: 18 July 2016; Accepted: 31 March 2017; Published: 30 April 2017. Nesting Ecology and Habitat Use of Chelydra serpentina in an Area Modified by Agricultural and Industrial Activity Molly M. Thompson1, Brittney H. Coe1, Justin D. Congdon2,3, Dean F. Stauffer1, and William A. Hopkins1,4 1Department of Fish and Wildlife Conservation, Virginia Tech, 106 Cheatham Hall, Blacksburg, Virginia 24062, USA 2Savannah River Ecological Laboratory, Drawer E, Aiken, South Carolina 29802, USA 3Bar Boot Ranch, Box 1128, Douglas, Arizona 85608, USA 4Corresponding author, email: [email protected] Abstract.—Habitat loss and pollution have been linked to declines of numerous freshwater turtle species, which are among the most endangered vertebrates in the world. We examined characteristics of nest sites of Common Snapping Turtles (Chelydra serpentina) located in a system modified by agricultural and industrial land use. We compared characteristics of 150 turtle nests and patterns of nest depredation in mercury (Hg) polluted and reference sites. Of the nests found in this study, 90% were located in human disturbed soils: 79% in agricultural fields and 11% in commercial nurseries. In both Hg and reference sites, we found that 52% of all nests were located in high density nesting areas in agricultural fields bordered by a river on three sides, providing novel evidence that river geomorphology could be useful for identifying important nesting areas. We did not observe predation in the reference sites but 66% of nests were destroyed at the Hg polluted sites. We provide a predictive model demonstrating that the same characteristics influence nest-site selection in this modified system as in more intact systems, and are related to solar exposure at the time of nesting. -

Tortugas (Testudinata) Marinas Y Continentales De Colombia
BiotaAcosta-Galvis Colombiana 1 (2) 187 - 194, 2000 Amphibians of Colombia - 187 Tortugas (Testudinata) Marinas y Continentales de Colombia Claudia Patricia Ceballos - Fonseca Department of Wildlife and Fisheries Sciences, Texas A&M University. [email protected] Palabras Clave: Tortugas, Testudinata, Taxonomía, Distribución, Colombia, Lista de Especies Durante los últimos trescientos años, diferentes cla- De Chelidae se hallan en Colombia tres géneros: Chelus, sificaciones naturales han sido propuestas para las tortu- Phrynops y Platemys. Del primero Chelus fimbriatus se gas. En la primera, presentada en 1766, Linnaeus agrupó las distribuye en las cuencas de los ríos Arauca, Orinoco y especies hasta entonces conocidas en el género Testudo. Amazonas. Del género Phrynops se cuentan cinco espe- Posteriormente, en 1805, Brongniart propuso una clasifica- cies: P. dahli, endémica de los ríos Sinú y San Jorge; P. ción con base en el hábitat por lo cual reconocía tres géne- geoffroanus, en los ríos Apaporis, Caquetá, Inírida, Muco ros: Chelonia para las tortugas marinas; Emys para las de y Vaupés; P. gibbus en el Caquetá y Meta; P. nasutus y P. agua dulce y Testudo para las terrestres. Nuevas propues- rufipes en el Amazonas y Vaupés. El género Platemys cuenta tas como las de Cuvier de 1817 y la de Wagler de 1830, con una sola especie: P. platycephala en el Amazonas, aunque aún emplearon el hábitat como criterio jerárquico, Caquetá, Putumayo y Orinoco (Medem 1961, 1968; King & se basaron en características anatómicas relacionadas con Burke 1989; Castaño 1992). aquel, como por ejemplo la presencia de membranas interdigitales en las tortugas de río (Gaffney 1984).