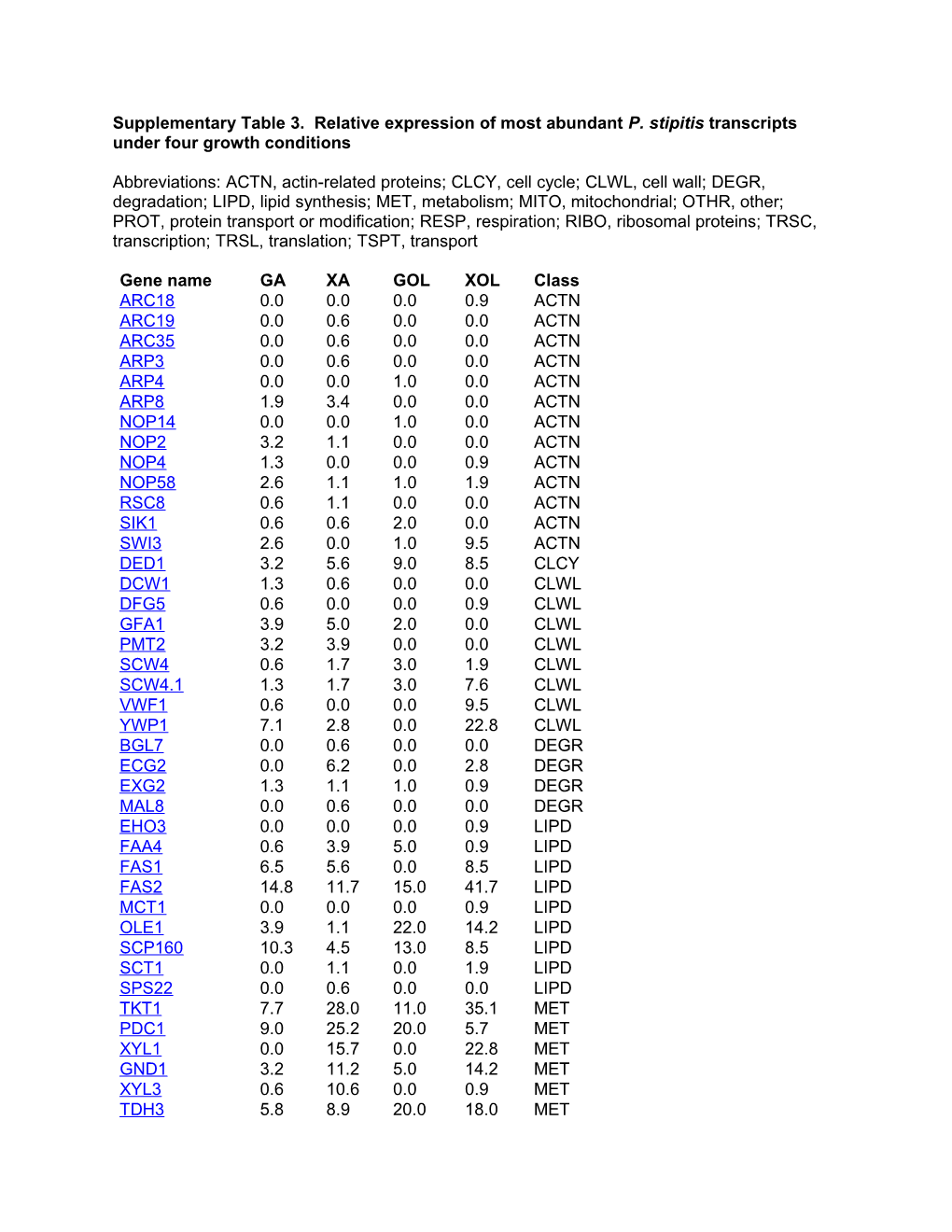
Supplementary Table 3. Relative expression of most abundant P. stipitis transcripts under four growth conditions
Abbreviations: ACTN, actin-related proteins; CLCY, cell cycle; CLWL, cell wall; DEGR, degradation; LIPD, lipid synthesis; MET, metabolism; MITO, mitochondrial; OTHR, other; PROT, protein transport or modification; RESP, respiration; RIBO, ribosomal proteins; TRSC, transcription; TRSL, translation; TSPT, transport
Gene name GA XA GOL XOL Class ARC18 0.0 0.0 0.0 0.9 ACTN ARC19 0.0 0.6 0.0 0.0 ACTN ARC35 0.0 0.6 0.0 0.0 ACTN ARP3 0.0 0.6 0.0 0.0 ACTN ARP4 0.0 0.0 1.0 0.0 ACTN ARP8 1.9 3.4 0.0 0.0 ACTN NOP14 0.0 0.0 1.0 0.0 ACTN NOP2 3.2 1.1 0.0 0.0 ACTN NOP4 1.3 0.0 0.0 0.9 ACTN NOP58 2.6 1.1 1.0 1.9 ACTN RSC8 0.6 1.1 0.0 0.0 ACTN SIK1 0.6 0.6 2.0 0.0 ACTN SWI3 2.6 0.0 1.0 9.5 ACTN DED1 3.2 5.6 9.0 8.5 CLCY DCW1 1.3 0.6 0.0 0.0 CLWL DFG5 0.6 0.0 0.0 0.9 CLWL GFA1 3.9 5.0 2.0 0.0 CLWL PMT2 3.2 3.9 0.0 0.0 CLWL SCW4 0.6 1.7 3.0 1.9 CLWL SCW4.1 1.3 1.7 3.0 7.6 CLWL VWF1 0.6 0.0 0.0 9.5 CLWL YWP1 7.1 2.8 0.0 22.8 CLWL BGL7 0.0 0.6 0.0 0.0 DEGR ECG2 0.0 6.2 0.0 2.8 DEGR EXG2 1.3 1.1 1.0 0.9 DEGR MAL8 0.0 0.6 0.0 0.0 DEGR EHO3 0.0 0.0 0.0 0.9 LIPD FAA4 0.6 3.9 5.0 0.9 LIPD FAS1 6.5 5.6 0.0 8.5 LIPD FAS2 14.8 11.7 15.0 41.7 LIPD MCT1 0.0 0.0 0.0 0.9 LIPD OLE1 3.9 1.1 22.0 14.2 LIPD SCP160 10.3 4.5 13.0 8.5 LIPD SCT1 0.0 1.1 0.0 1.9 LIPD SPS22 0.0 0.6 0.0 0.0 LIPD TKT1 7.7 28.0 11.0 35.1 MET PDC1 9.0 25.2 20.0 5.7 MET XYL1 0.0 15.7 0.0 22.8 MET GND1 3.2 11.2 5.0 14.2 MET XYL3 0.6 10.6 0.0 0.9 MET TDH3 5.8 8.9 20.0 18.0 MET XYL2 0.0 7.8 0.0 12.3 MET ACS2 3.9 4.5 2.0 4.7 MET FBA1 0.6 3.4 10.0 0.9 MET TAL1 1.9 3.4 5.0 11.4 MET TSA1 0.0 3.4 0.0 0.0 MET ADH1 4.5 2.8 4.0 0.9 MET GLK1 0.6 2.8 1.0 0.0 MET LAT1 1.3 2.8 4.0 0.9 MET PGM2 0.0 2.8 2.0 2.8 MET PGK1 19.4 2.2 5.0 3.8 MET ARD2 0.0 1.7 4.0 1.9 MET GDH2 1.9 1.7 2.0 13.3 MET GPM1.1 0.0 1.7 4.0 4.7 MET PDA1 1.3 1.7 3.0 1.9 MET PYC1 7.1 1.7 2.0 7.6 MET PGI1 2.6 1.1 3.0 6.6 MET SCT1 0.0 1.1 0.0 1.9 MET SOL2 0.0 1.1 0.0 0.0 MET TPI1 0.0 1.1 2.0 1.9 MET ADH3 0.0 0.6 0.0 0.0 MET ENO1 3.9 0.6 3.0 5.7 MET FBA1 0.0 0.6 0.0 0.0 MET FBP1 0.0 0.6 1.0 0.0 MET GLK2 0.0 0.6 0.0 0.0 MET GLN1 1.3 0.6 6.0 5.7 MET GLN4 1.3 0.6 6.0 0.0 MET GLT1 1.3 0.6 0.0 0.9 MET PDB1 1.3 0.6 3.0 0.9 MET PFK1 1.3 0.6 2.0 3.8 MET RBK1 0.0 0.6 0.0 0.0 MET RKS1 0.6 0.6 6.0 0.0 MET SOU2 0.0 0.6 6.0 4.7 MET TDH2 3.2 0.6 3.0 1.9 MET ZWF1 1.3 0.6 0.0 0.0 MET ACS1 0.6 0.0 0.0 0.0 MET ALD1 0.6 0.0 1.0 0.0 MET GDH3 0.0 0.0 1.0 0.9 MET HXK1 0.6 0.0 0.0 0.0 MET KPR5 0.0 0.0 0.0 0.9 MET PCK1 0.0 0.0 1.0 0.0 MET PDC2 0.0 0.0 0.0 0.9 MET PDX1 0.0 0.0 0.0 0.9 MET PFK2 1.3 0.0 0.0 1.9 MET PGM3 0.6 0.0 0.0 0.0 MET PRS2 0.6 0.0 0.0 0.0 MET PRS3 0.0 0.0 1.0 0.0 MET PYK1 0.0 0.0 2.0 1.9 MET RKI1 0.0 0.0 0.0 0.9 MET SAD2 0.0 0.0 1.0 0.9 MET TDH1 0.0 0.0 2.0 0.0 MET ACO1 17.4 10.6 4.0 10.4 MITO ADT1 5.2 15.1 6.0 6.6 MITO CIT1 3.2 0.6 1.0 1.9 MITO DLD2 0.0 0.6 1.0 0.0 MITO FUM1 0.0 1.1 0.0 0.9 MITO IDH1 0.0 0.6 1.0 2.8 MITO IDH2 1.3 1.1 0.0 2.8 MITO IDP1 0.0 0.6 0.0 0.0 MITO ILV2 0.0 0.6 2.0 2.8 MITO KDG2 7.1 2.2 6.0 1.9 MITO KGD1 10.3 2.8 2.0 1.9 MITO LPD1 1.3 1.1 2.0 0.0 MITO MAE1 4.5 14.0 0.0 0.0 MITO MDH1 1.3 2.2 1.0 2.8 MITO MDH2 0.6 0.0 0.0 0.0 MITO MIR1 0.0 3.9 1.0 1.9 MITO SDH1 3.2 3.9 2.0 7.6 MITO SDH2 0.0 0.6 0.0 0.0 MITO SDH4 0.6 0.6 0.0 0.9 MITO SDH6 1.3 0.0 0.0 1.9 MITO ADE6 5.2 2.2 1.0 8.5 OTHR CAN2 0.0 0.0 7.0 0.0 OTHR GPH1 2.6 1.7 7.0 8.5 OTHR IDI1 0.6 0.0 0.0 0.0 OTHR MET6 6.5 2.8 8.0 4.7 OTHR PAN1 3.2 3.4 14.0 5.7 OTHR SSN6 0.0 0.0 9.0 2.8 OTHR TCT1 1.9 1.1 7.0 1.9 OTHR TSA1 1.9 2.2 1.0 0.0 OTHR KIN2 2.6 1.1 10.0 2.8 PROT PHO8 2.6 1.7 8.0 7.6 PROT SEC31 7.1 5.6 18.0 3.8 PROT SEC61 1.9 1.1 2.0 0.9 PROT VTC1 1.9 0.0 0.0 0.0 PROT VTC2 0.0 2.2 0.0 0.0 PROT VTC4 5.8 2.8 0.0 0.0 PROT AOX1 0.6 2.8 7.0 0.0 RESP COX11 0.6 0.0 0.0 0.0 RESP COX12 0.0 0.0 0.0 1.9 RESP COX13 0.0 0.6 0.0 0.9 RESP COX15 0.0 0.6 1.0 0.0 RESP COX17 0.0 0.0 1.0 0.0 RESP COX18 0.0 0.6 0.0 0.0 RESP COX4 0.0 0.6 1.0 0.0 RESP COX5a 0.0 2.2 0.0 0.0 RESP COX6 0.0 1.1 0.0 2.8 RESP COX7 0.0 0.6 1.0 0.0 RESP CUP5 0.6 0.6 0.0 0.0 RESP CYB2 0.0 0.6 0.0 0.0 RESP CYC1 2.6 1.7 0.0 1.9 RESP CYT1 0.0 1.1 1.0 0.0 RESP NDE1 3.2 1.7 3.0 2.8 RESP NUO78 5.2 2.2 1.0 0.0 RESP TFP1 4.5 1.1 1.0 0.0 RESP TFP3 0.6 0.0 0.0 0.0 RESP VPH1 0.6 2.2 0.0 0.0 RESP VPH3 0.6 0.6 0.0 0.0 RESP CLG1 0.0 0.0 2.0 5.7 RIBO Hypothetical 0.0 9.5 0.0 0.0 RIBO protein RPL15A 2.6 11.7 1.0 2.8 RIBO RPL16B 3.2 5.0 5.0 9.5 RIBO RPL2A 1.9 0.0 0.0 4.7 RIBO RPL2B 2.6 0.0 0.0 0.0 RIBO RPL3 5.2 9.5 8.0 3.8 RIBO RPP0 5.2 9.5 2.0 5.7 RIBO RPS20 5.2 10.6 0.0 1.9 RIBO RPS7A 6.5 6.2 3.0 0.0 RIBO FCR1 0.0 0.6 0.0 0.0 TRSC FST1 0.6 0.0 0.0 0.9 TRSC FST10 0.0 0.6 2.0 0.0 TRSC FST11 0.0 0.6 0.0 0.9 TRSC FST13 0.6 0.0 0.0 1.9 TRSC FST5 0.6 0.0 0.0 1.9 TRSC GIN1 0.0 0.0 4.0 0.0 TRSC HBR1 0.0 0.6 0.0 0.0 TRSC MBT1 0.0 0.6 0.0 0.0 TRSC NAF2 0.0 0.6 0.0 0.0 TRSC RGR1 0.6 0.0 0.0 0.9 TRSC SFP1 0.6 0.0 6.0 0.0 TRSC SIN3 1.3 2.2 13.0 5.7 TRSC STB4 0.6 0.6 1.0 0.0 TRSC STD1 0.0 0.0 0.0 0.9 TRSC SUC1.1 0.6 0.0 0.0 0.0 TRSC SUC1.3 0.6 0.6 0.0 0.0 TRSC WAR1 0.6 0.0 1.0 0.9 TRSC YIN0 0.6 0.6 1.0 0.9 TRSC IF5A 6.5 5.0 0.0 0.9 TRSL NIP1 5.2 3.4 3.0 2.8 TRSL PAB1 11.0 5.0 2.0 12.3 TRSL TIF1 1.3 3.4 0.0 0.9 TRSL TIF4631 2.6 0.0 41.0 6.6 TRSL DIP5.3 0.6 1.7 6.0 3.8 TSPT ENA2 1.3 0.0 0.0 0.0 TSPT ENA5 25.8 4.5 8.0 2.8 TSPT HXT2.4 0.0 0.6 0.0 0.0 TSPT PHO84 5.8 1.7 31.0 24.7 TSPT PHO89 7.7 2.2 6.0 0.0 TSPT PMA1 0.0 0.6 0.0 0.9 TSPT PMC1 5.8 3.9 31.0 19.0 TSPT QUP1 0.0 0.0 1.0 0.0 TSPT RGT2 0.0 1.7 2.0 1.9 TSPT SSY1.5 0.6 0.6 2.0 0.0 TSPT SUT1 3.2 0.6 1.0 0.0 TSPT SUT4 0.6 2.2 0.0 0.0 TSPT