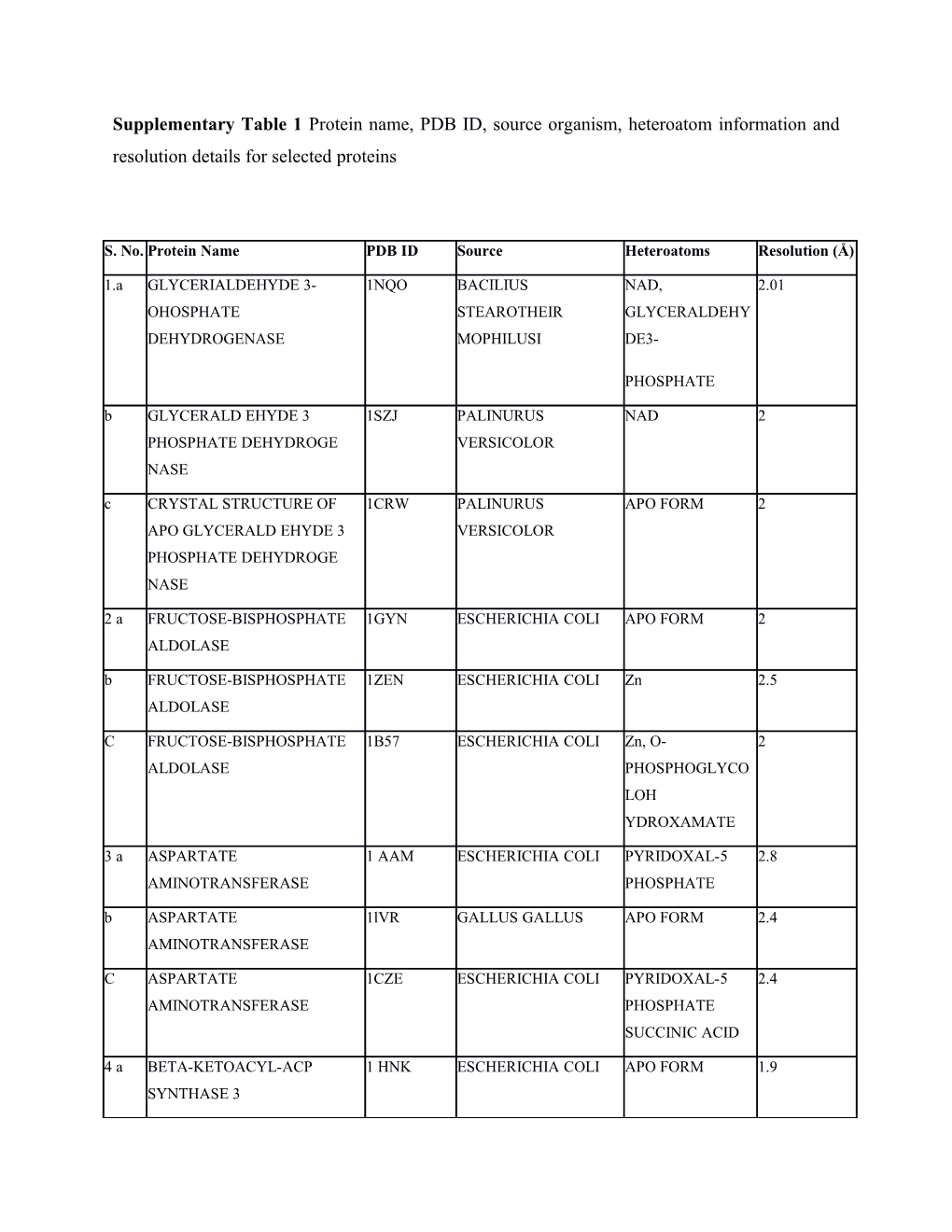
Supplementary Table 1 Protein name, PDB ID, source organism, heteroatom information and resolution details for selected proteins
S. No. Protein Name PDB ID Source Heteroatoms Resolution (Å)
1.a GLYCERIALDEHYDE 3- 1NQO BACILIUS NAD, 2.01 OHOSPHATE STEAROTHEIR GLYCERALDEHY DEHYDROGENASE MOPHILUSI DE3-
PHOSPHATE b GLYCERALD EHYDE 3 1SZJ PALINURUS NAD 2 PHOSPHATE DEHYDROGE VERSICOLOR NASE c CRYSTAL STRUCTURE OF 1CRW PALINURUS APO FORM 2 APO GLYCERALD EHYDE 3 VERSICOLOR PHOSPHATE DEHYDROGE NASE
2 a FRUCTOSE-BISPHOSPHATE 1GYN ESCHERICHIA COLI APO FORM 2 ALDOLASE b FRUCTOSE-BISPHOSPHATE 1ZEN ESCHERICHIA COLI Zn 2.5 ALDOLASE
C FRUCTOSE-BISPHOSPHATE 1B57 ESCHERICHIA COLI Zn, O- 2 ALDOLASE PHOSPHOGLYCO LOH YDROXAMATE
3 a ASPARTATE 1 AAM ESCHERICHIA COLI PYRIDOXAL-5 2.8 AMINOTRANSFERASE PHOSPHATE b ASPARTATE 1lVR GALLUS GALLUS APO FORM 2.4 AMINOTRANSFERASE
C ASPARTATE 1CZE ESCHERICHIA COLI PYRIDOXAL-5 2.4 AMINOTRANSFERASE PHOSPHATE SUCCINIC ACID
4 a BETA-KETOACYL-ACP 1 HNK ESCHERICHIA COLI APO FORM 1.9 SYNTHASE 3 b BETA-KETOACYL-ACP 1EBL ESCHERICHIA COLI SELENOMETHION 1.8 SYNTHASE 3 ION
C BETA-KETOACYL-ACP 1 HND ESCHERICHIA COLI COENZYME A 1.6 SYNTHASE 3
5 a LACATATE 6LDH SQUALUS ACANTHIAS APO FORM 2 DEHYDROGENASE b LACATATE 1LLD BIFIDOBACTER IUM NAD 2 DEHYDROGENASE LONGUM c LACATATE 1U5C PLASMODIUM NAD, 2.65 DEHYDROGENASE FALCIPARUM DIHYDROXY-2- NAPHTHOIC ACID
6 a THYMIDYLATE SYNTHASE 1 TIS BACTERIOPHA GE APO FORM 3.1 b THYMIDYLATE SYNTHASE 2 BBQ ESCHERICHIA COLI 2’- 2.3 DEOXYURIDINE5, MONOPHOSPHAT E10- PARPARGYL-5-8 DIDEAZAFOLATE -4- GLUTAMIC ACID c THYMIDYLATE SYNTHASE 1 DNA ESCHERICHIA COLI 2’- 2.2 DEOXYURIDINE5, MONOPHOSPHAT E
7 a GLUCODE DEHYDROGENASE 1CQ1 ACINETOBACTER PYRROLOQUINO 1.9 CALCOACETICUS LINE QUINONE GLUCOSE b GLUCODE DEHYDROGENASE 1C9U ACINETOBACTER PYRROLOQUINO 2.2 CALCOACETICUS LINE QUINONE c GLUCODE DEHYDROGENASE 1QBI ACINETOBACTER APO FORM 1.72 CALCOACETICUS
8 a ALDEHYDE 1EUH STREPTOCOCCUS APO FORM 1.82 DEHYDROGENASE MUTANS b ALDEHYDE 2EUH STREPTOCOCCUS NADP 2.6 DEHYDROGENASE MUTANS c ALDEHYDE 1QI1 STREPTOCOCCUS NADP AND D- 3 DEHYDROGENASE MUTANS GLYCERALDEHY DE-3 PHOSPHATE
9 a ADENOSINE DEAMINASE 1ADD ESCHERICHIA COLI Zn, ADINOSINE 2.4
ADENOSINE DEAMINASE 1VFL BOS TAURUS Zn 1.8
ADENOSINE DEAMINASE 2AMX PLASMODIUM YOELII APO FORM 2.02
10 a GLUTATHIONE REDUCTASE 1GRA HOMO SAPIENS GLUTATHIONE 2 NADPH b GLUTATHIONE REDUCTASE 1GRT ESCHERICHIA COLI APO 2.3 FORM(PHYSIOLO GICALY PRESENT WITH FAD) c GLUTATHIONE REDUCTASE 1GET ESCHERICHIA COLI NADPH 2
11 a COPPER-CONTAINING AMINE 1IU7 AROTHROBACTEE Cu, TPQ 1.8 OXIDASE GLOBIFORMIS b AMINE OXIDASE 1WMP AROTHROBACTEE APO FORM 2 GLOBIFORMIS c COPPER-CONTAINING AMINE 1W5Z ESCHERICHIA COLI Cu, TPQ, 1.86 OXIDASE BENZYLHYDRAZI NE
12 a BILIVERDIN REDUCTASE 1GCU RATTUS NORVEGICUS APO FORM 1.4 b BILIVERDIN REDUCTASE 1HE2 HOMOSAPIENS NADP, 1.2 BILIVERDINEIXA LPHA c BILIVERDIN REDUCTASE 1LC3 RATTUS NORVEGICUS NADP, 1.5
13 1NU EMERICELLA APO FORM 2.85 a 3-DEHYDROQUINATE A NIDULANS SYNTHASE b 3-DEHYDROQUINATE 1NRX EMERICELLA NAD 2.9 SYNTHASE NIDULANS
C 3-DEHYDROQUINATE 1NVB EMERICELLA NAD 2.7 SYNTHASE NIDULANS CARBAPHOSPHO NATE
14 a MALIC ENZYME 2AW5 HOMO SAPIENS APO FORM 2.5 b MALIC ENZYME 1LLQ ASCARIS SUUM NAD 2.3 c MALIC ENZYME 1EFK HOMO SAPIENS NAD ALPHA- 2.6 KETOMALONIC ACIO
15 a MYROSINASE 1.00E+72 SINAPIS ALBA Cofactor, GLUCO 1.6 HYDROXIMOLAC TAM b MYROSINASE 1.00E+71 SINAPIS ALBA ASCROBIC ACID 1.5 (COFACTOR) c MYROSINASE 1MYR SINAPIS ALBA APO FORM 1.64