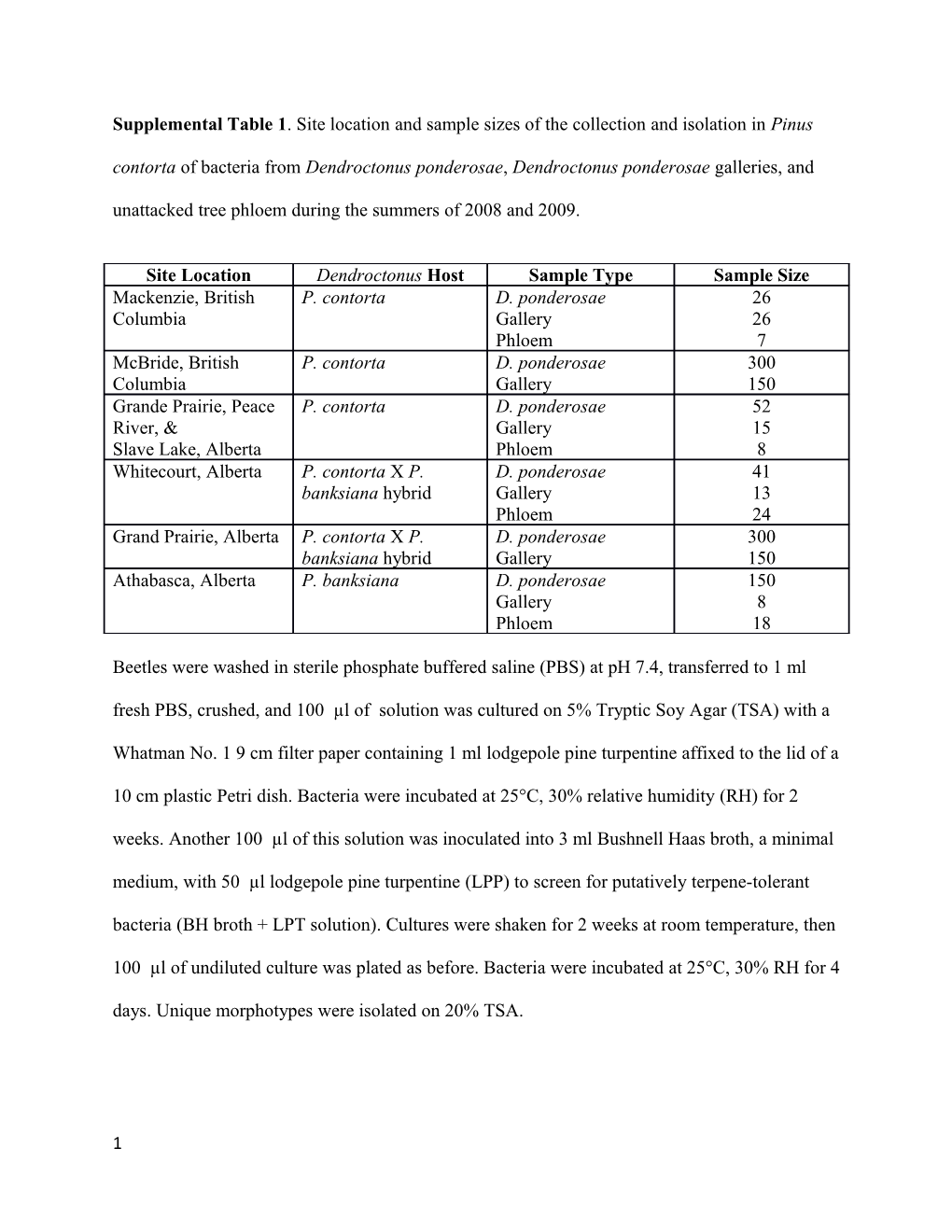
Supplemental Table 1. Site location and sample sizes of the collection and isolation in Pinus contorta of bacteria from Dendroctonus ponderosae, Dendroctonus ponderosae galleries, and unattacked tree phloem during the summers of 2008 and 2009.
Site Location Dendroctonus Host Sample Type Sample Size Mackenzie, British P. contorta D. ponderosae 26 Columbia Gallery 26 Phloem 7 McBride, British P. contorta D. ponderosae 300 Columbia Gallery 150 Grande Prairie, Peace P. contorta D. ponderosae 52 River, & Gallery 15 Slave Lake, Alberta Phloem 8 Whitecourt, Alberta P. contorta X P. D. ponderosae 41 banksiana hybrid Gallery 13 Phloem 24 Grand Prairie, Alberta P. contorta X P. D. ponderosae 300 banksiana hybrid Gallery 150 Athabasca, Alberta P. banksiana D. ponderosae 150 Gallery 8 Phloem 18
Beetles were washed in sterile phosphate buffered saline (PBS) at pH 7.4, transferred to 1 ml fresh PBS, crushed, and 100 µl of solution was cultured on 5% Tryptic Soy Agar (TSA) with a
Whatman No. 1 9 cm filter paper containing 1 ml lodgepole pine turpentine affixed to the lid of a
10 cm plastic Petri dish. Bacteria were incubated at 25°C, 30% relative humidity (RH) for 2 weeks. Another 100 µl of this solution was inoculated into 3 ml Bushnell Haas broth, a minimal medium, with 50 µl lodgepole pine turpentine (LPP) to screen for putatively terpene-tolerant bacteria (BH broth + LPT solution). Cultures were shaken for 2 weeks at room temperature, then
100 µl of undiluted culture was plated as before. Bacteria were incubated at 25°C, 30% RH for 4 days. Unique morphotypes were isolated on 20% TSA.
1 Supplemental Table 2. Bacteria isolated from Dendroctonus ponderosae adults and assayed for potential ability to metabolize
terpenes.
Isolate (Accession Closest named match Sequence Length S_ab Score number) (Acc. No.) (base pairs) JN194137 Serratia marcescens (AB061685) 1414 0.975 JN194138 Brevundimonas vesicularis (AJ227780) 1320 0.985 JN194126 Serratia marcescens (AB061685) 1394 0.984 JN194139 Pseudomonas brenneri (AF268968) 1392 0.975 JN194140 Pseudomonas mandelii (AF058286) 952 0.978 JN194133 Pantoea agglomerans (AJ233423) 1289 0.921 JN194135 Rahnella aquatilis (AJ233426) 1398 0.917 JN194127 Pseudomonas migulae (AF074383) 1386 0.973
Target sequences were amplified by performing a polymerase chain reaction (PCR) using the primers 27f (5’-
GAGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’). Sequencing of PCR products was performed by the University of Wisconsin Biotechnology Center. Sequence data were aligned using Sequencher (version 4.4 build
1415, Gene Codes Corporation, Ann Arbor, MI). Sequences of bacteria used in bioassays were deposited in GenBank. The closest match, S_ab score and accession number of the closest match were obtained using the Ribosomal Database Project
(http://rdp.cme.msu.edu/) by searching for type strain, source isolates, ≥1200 base pairs in size and good quality.
2 Supplemental Table 3. Details of materials and procedures used in bioassays. Listed in order of presentation in text.
Material Vendor Details Tryptic Soy Broth MP Biomedicals, Solon, OH, USA Lodgepole pine Synergy Semiochemicals turpentine Corp, Burnaby, BC, Canada Filter paper Whatman TSA Agar; Difco, Sparks, MD, USA PTFE thread tape seal AA Thread Seal Tape Co., 1.3 cm wide Wauconda, IL, USA Clear screw-cap glass Fisher Scientific, Pittsburg, 25 X 83mm 6 dram vials PA, USA Monoterpenes Aldrich, St, Louis, MO, USA α-pinene [98%; blend: (-):(+)-α- pinene: 44:54]; (-)-β-pinene (>88%), 3-carene (90%) Resolv hexanes Fisher Scientific, Pittsburg, PA, USA Autosampler vials National Scientific isobutylbenzene Aldrich, St. Louis, MO, USA 98% Gas chromatography Shimadzu 17-A GC-FID Temperature cycle: 10 minutes at 60˚C, then 5˚C min-1 to 160˚C. Injectors and detectors at 220˚C. Chiral column Agilent Technologies Cyclodex-B; 30 m, 0.25 mm i.d., 0.25 µm film thickness Kruskal-Wallis Test Systat Software, Point SYSTAT 11 Richmond, CA, USA
Media for abiotic acid 0.3 g/L yeast extract, 6.0 g/L Na2HPO4, 3.0 g/L KH2PO4, 0.5 g/L assays NaCl, 1.0 g NH4Cl, 1.0g KNO3, 1M MgSO4 Vacuum centrifuge Savant Speedvac High-pressure liquid Hewlett Packard; 1090 with Mobile phase consisted of chromatography diode array detector (DAD); MeOH:1.7% acetic acid (91:9) run ODS 5 µm 4.6 x 25 mm isocratically at 1.0 ml/min with column; Beckman Ultrasphere column heater at 30C. We injected 20 µl of each solution and compared retention times and peak areas at 240 nm to abietic acid standard. We collected UV data from 190-400 nm and confirmed identity in each sample with resulting spectra. All peak areas fell within linear range (R2 = 1.0, P < 0.001) of standard. Abietic acid Acros Organics, Waltham, 85% MA, USA 3 ANOVA SAS Institute, Cary NC, USA SAS version 9.1, 2003; PROC GLM Petri dishes Fisher Scientific, Pittsburg, PA, USA Yeast Malt Extract 4 g/L yeast extract, 10 g/l malt extract broth, 4 g/l dextrose, 20 g/l agar; Agar Spores were dislodged from hyphae using a bacterial spreader.
4