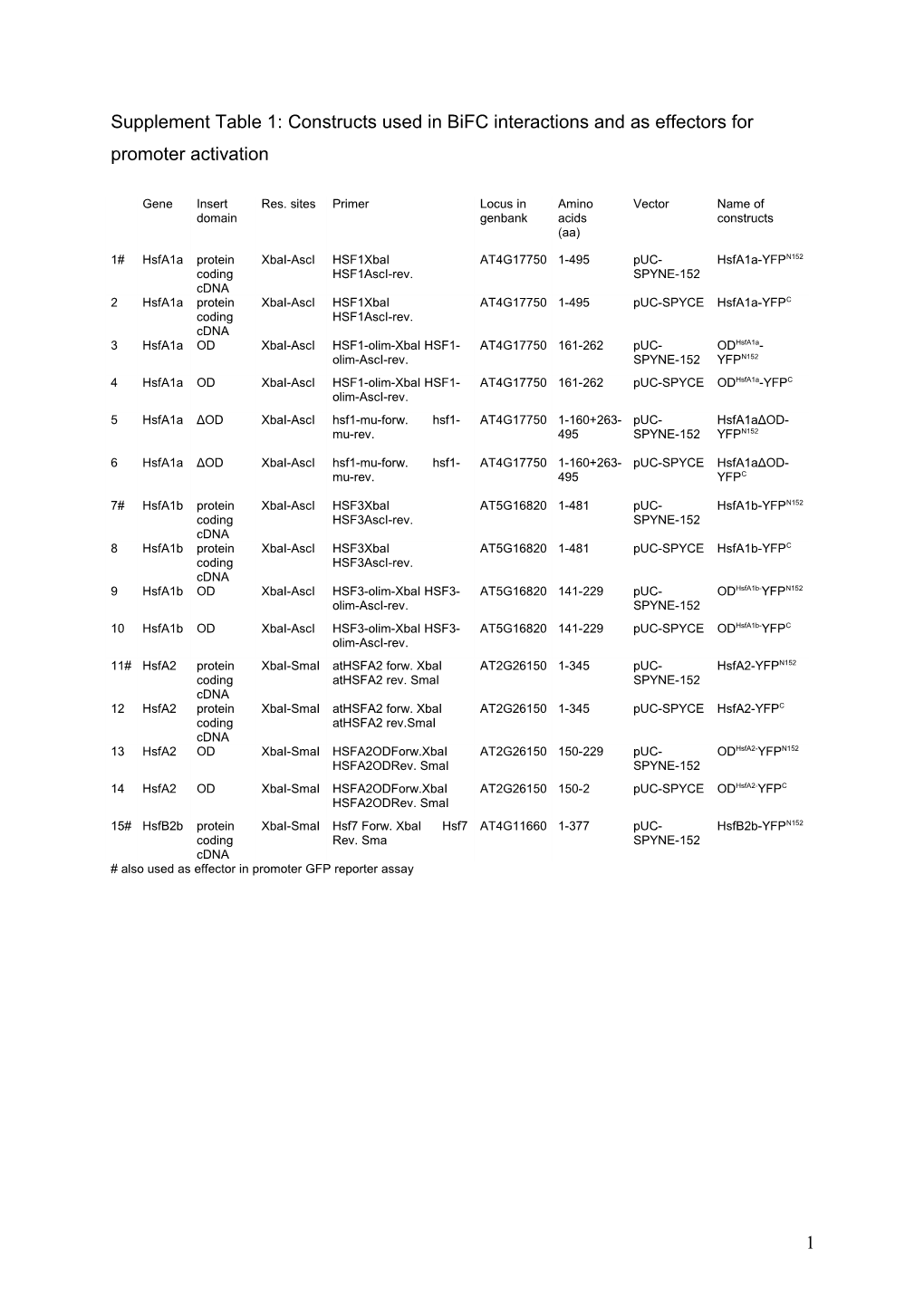
Supplement Table 1: Constructs used in BiFC interactions and as effectors for promoter activation
Gene Insert Res. sites Primer Locus in Amino Vector Name of domain genbank acids constructs (aa)
1# HsfA1a protein XbaI-AscI HSF1XbaI AT4G17750 1-495 pUC- HsfA1a-YFPN152 coding HSF1AscI-rev. SPYNE-152 cDNA 2 HsfA1a protein XbaI-AscI HSF1XbaI AT4G17750 1-495 pUC-SPYCE HsfA1a-YFPC coding HSF1AscI-rev. cDNA 3 HsfA1a OD XbaI-AscI HSF1-olim-XbaI HSF1- AT4G17750 161-262 pUC- ODHsfA1a- olim-AscI-rev. SPYNE-152 YFPN152 4 HsfA1a OD XbaI-AscI HSF1-olim-XbaI HSF1- AT4G17750 161-262 pUC-SPYCE ODHsfA1a-YFPC olim-AscI-rev. 5 HsfA1a ΔOD XbaI-AscI hsf1-mu-forw. hsf1- AT4G17750 1-160+263- pUC- HsfA1aΔOD- mu-rev. 495 SPYNE-152 YFPN152
6 HsfA1a ΔOD XbaI-AscI hsf1-mu-forw. hsf1- AT4G17750 1-160+263- pUC-SPYCE HsfA1aΔOD- mu-rev. 495 YFPC
7# HsfA1b protein XbaI-AscI HSF3XbaI AT5G16820 1-481 pUC- HsfA1b-YFPN152 coding HSF3AscI-rev. SPYNE-152 cDNA 8 HsfA1b protein XbaI-AscI HSF3XbaI AT5G16820 1-481 pUC-SPYCE HsfA1b-YFPC coding HSF3AscI-rev. cDNA 9 HsfA1b OD XbaI-AscI HSF3-olim-XbaI HSF3- AT5G16820 141-229 pUC- ODHsfA1b-YFPN152 olim-AscI-rev. SPYNE-152 10 HsfA1b OD XbaI-AscI HSF3-olim-XbaI HSF3- AT5G16820 141-229 pUC-SPYCE ODHsfA1b-YFPC olim-AscI-rev. 11# HsfA2 protein XbaI-SmaI atHSFA2 forw. XbaI AT2G26150 1-345 pUC- HsfA2-YFPN152 coding atHSFA2 rev. SmaI SPYNE-152 cDNA 12 HsfA2 protein XbaI-SmaI atHSFA2 forw. XbaI AT2G26150 1-345 pUC-SPYCE HsfA2-YFPC coding atHSFA2 rev.SmaI cDNA 13 HsfA2 OD XbaI-SmaI HSFA2ODForw.XbaI AT2G26150 150-229 pUC- ODHsfA2-YFPN152 HSFA2ODRev. SmaI SPYNE-152 14 HsfA2 OD XbaI-SmaI HSFA2ODForw.XbaI AT2G26150 150-2 pUC-SPYCE ODHsfA2-YFPC HSFA2ODRev. SmaI 15# HsfB2b protein XbaI-SmaI Hsf7 Forw. XbaI Hsf7 AT4G11660 1-377 pUC- HsfB2b-YFPN152 coding Rev. Sma SPYNE-152 cDNA # also used as effector in promoter GFP reporter assay
1 Supplement Table 2: HSF constructs used in Yeast 2-hybrid interactions
Gene Insert Res. Primer Locus in Amino Vector Name of site genbank acids constructs (aa)
1 HsfA1a protein coding SmaI Hsf1 SmaI Forw.2 AT4G17750 2-496 pGADT7 AD-HsfA1a cDNA Hsf1 SmaI Rev.2 2 HsfA1a protein coding SmaI Hsf1 SmaI Forw.2 AT4G17750 2-496 pGBKT7 BD-HsfA1a cDNA Hsf1 SmaI Rev.2 3 HsfA1b protein coding EcoRI Hsf3 EcoRI Forw. AT5G16820 2-482 pGADT7 AD-HsfA1b cDNA Hsf3 EcoRI Rev.
4 HsfA1b protein coding EcoRI Hsf3 EcoRI Forw. AT5G16820 2-482 pGBKT7 BD-HsfA1b cDNA Hsf3 EcoRI Rev. 5 HsfA2 protein coding SmaI hsfA2 SmaI Forw. AT2G26150 2-346 pGADT7 AD-HsfA2 cDNA hsfA2 SmaI Rev. 6 HsfA2 protein coding SmaI hsfA2 SmaI Forw. AT2G26150 2-346 pGBKT7 BD-HsfA2 cDNA hsfA2 SmaI Rev.
2 Supplement Table 3: Promoter GFP reporter constructs used in activation assays
Gene Insert Insert Primer Locus in Length of Vector Name of constructs enzyme genbank upstream sequence (bp)* 1 pHSP26.5 5´upstream BamHI Hsp26.5-P Forw AT1g52560 1680 pGTkan pHsp26.5-GFP Hsp26.5-P Rev 2 pHSP18.1-CI 5´upstream SacI AT5G59720 895 pGTkan pHsp18.1-CI GFP
3 pHSP21 5´upstream BamHI Hsp21 Forw AT4G27670 1367 pGTkan pHsp25.3-GFP Hsp21 Rev 4 pHSP17.6C- 5´upstream BamHI Hsp17.6C-CI AT1G53540 731 pGTkan pHsp17.6C-CI-GFP CI Forw Hsp17.6C-CI Rev 5 pHSP17.6CII 5´upstream BamHI Hsp17.6-CII AT5G12020 1337 pGTkan pHsp17.6CII-GFP Forw Hsp17.6CII Rev 6 pHsfA2 5´upstream BamHI HSFA2 Forw AT2G26150 729 pGTkan pHsfA2-GFP HSFA2 Rev *)Sequences starting immediately upstream of the ATG of the protein coding region
3 Supplement Table 4: Primers used in Hsf and reporter gene constructions
Restriction Name Forward (F) and reverse (R) primer sequences Length enzyme HSF1XbaI F 5'-CTGCTCTAGAGCATGTTTGTAAATTTCAAATACTTCTCTTTC-3' 42 XbaI
HSF1AscI-rev. R 5'-TTGGCGCGCCTGTGTTCTGTTTCTGATGTGAGAAG-3' 35 AscI
HSF1-olim-XbaI F 5'-CTGCTCTAGAGCATGTCTCAGGGTCAAGGTTC-3' 32 XbaI
HSF1-olim-AscI- R 5'-TTGGCGCGCCTATTGGCCTCGGTTACATG-3' 29 AscI rev. hsf1-mu-forw. F 5'-CATGACAAGAAGCGGAGACTCAGAGAG-3' 27 -
hsf1-mu-rev. R 5'-GTACTGTAATTGCTGAGATTGTGGATTACTAC-3' 31 -
Hsf1 SmaI Forw.2 F 5'-TCCCCCGGGGGGATTTGTAAATTTCAAATACTTCTCTTTC-3' 40 SmaI
Hsf1 SmaI Rev.2 R 5'-TCCCCCGGGGGGACTAGTGTTCTGTTTCTGATGTGAG-3' 37 SmaI
HSF3XbaI F 5'-CTGCTCTAGAGCATGGAATCGGTTCCCG-3' 28 XbaI
HSF3AscI-rev. R 5'-TTGGCGCGCCTTTTCCTCTGTGCTTCTGAG-3' 30 AscI
HSF3-olim-XbaI F 5'-CTGCTCTAGAGCATGGAGGTGGGGAAGTTTG-3' 31 XbaI
HSF3-olim-AscI- R 5'-TTGGCGCGCCTGTTGCTTCCTGGAATTTGTCTG-3' 33 AscI rev. Hsf3 EcoRI Forw. F 5'-CCGGAATTCCGGGAATCGGTTCCCGAATC-3' 29 EcoRI
Hsf3 EcoRI Rev. R 5'-CCGGAATTCCGGTTATTTCCTCTGTGCTTCTGAG-3' 34 EcoRI
atHSFA2 forw.XbaI F 5'-GCTCTAGAGCATGGAAGAACTGAAAGTGGAAA-3' 32 XbaI
atHSFA2 rev. SmaI R 5'-TCCCCCGGGGGAAGGTTCCGAACCAAGAAAAC-3' 32 SmaI HSFA2 OD F 5'-GCTCTAGAGCATGTCATGTGTTGAGGTTGGG-3' 31 XbaI Forw.XbaI HSFA2 OD Rev. R 5'-TCCCCCGGGGGAACTCATAACCGCAAACTGCT-3' 32 SmaI SmaI hsfA2 SmaI Forw. F 5'-TCCCCCGGGGGGAGAAGAACTGAAAGTGGAAATGG-3' 35 SmaI
hsfA2 SmaI Rev. R 5'-TCCCCCGGGGGATTAAGGTTCCGAACCAAGAA-3' 32 SmaI Hsf7 Forw. XbaI F 5'-GCTCTAGAGCATGCCGGGGGAACAA-3' 25 XbaI
Hsf7 Rev. SmaI R 5'-TCCCCCGGGGGATTTTCCGAGTTCAAGCCAC-3' 31 SmaI
Hsp26.5-P Forw F 5'-CGCGGATCCGCGCATCATCATGCTGTGGTTTATAT-3' 35 BamHI
Hsp26.5-P Rev R 5'-CGCGGATCCGCGTGTTTTCAAATCGGTAAATTTCTTA-3' 37 BamHI
Hsp21 Forw F 5'-CGCGGATCCGCGTCTCTTCTAAAAATTCAACCTTTC-3' 36 BamHI
Hsp21 Rev R 5'-CGCGGATCCGCGTTGTTTCGAGTATGAGCCA-3' 31 BamHI
Hsp17.6C-CI Forw F 5'-CGCGGATCCGCGATTATATACACATACACATTCAGG-3' 36 BamHI
Hsp17.6C-CI Rev R 5'-CGCGGATCCGCGTTTCACTTCCTCTTGTG-3' 29 BamHI
Hsp17.6-CII Forw F 5'-CGCGGATCCGCGTTTGTTTGTGATCGTGTTTT-3' 32 BamHI
Hsp17.6CII Rev R 5'-CGCGGATCCGCGTGTTAGTTGTGTTGTGTTTGC-3' 33 BamHI
HSFA2 Forw F 5'-CGCGGATCCGCGCAATCAAATCTCTCCTTCACG-3' 33 BamHI
HSFA2 Rev R 5'-CGCGGATCCGCGTTTCGTTGTTTATCTCAAATCCA-3' 35 BamHI
4 Supplement Figures S1-S3
Figure S1: Western blot immunodetection of expression levels of recombinant HsfA1a and HsfA1b in protoplasts in the promoter activation assay (corresponding with the data depicted in Figure 3). The protein samples were isolated from the same transformed protoplasts as used in one of the promoter activation assays (promoter-GFP constructs tested: pHsp17.6C- CI, pHsp17.6CII, pHsfA2, pHsp26.5-P(r),pHsp 18.1-CI, pHsp-25.3-P) contributing to the results shown in Fig. 3 of the manuscript. Hsf expression was detected using Myc-antibody. Transformed effectors: 1) bZIP63; 2) HsfA1a; 3) HsfA1b; 4) HsfA1a/HsfA1b.
Figure S2: Western blot immunodetection of expression levels of HsfA1, HsfA1b and HsfA2 in transformed protoplasts in the promoter activation assay (corresponding with the data depicted in Figure 4). The protein samples were isolated from the same transformed protoplasts as used in one of the promoter activation assays (Promoter: pHsp 18.1-CI) by early and (HsfA1a, Hsf A1b) late (HsfA2) class A Hsfs. Hsf expression was detected using Myc-antibody. Transformed effectors 1) bZIP63; 2) HsfA1a; 3) HsfA2; 4) HsfA1a/A2; 5) HsfA1b/A2.
5 Figure S3: System test of promoter activation assay - activation of the pHSP25.6-P-GFP reporter by different effectors. Arabidopsis protoplasts were transformed with the promoter pHSP25.6-P-GFP reporter plasmids in combinations different DNAs tested as effectors. Plasmids (25 µg) expressing the effectors YFPN152 (expressing only the N-terminal part of YFP), HsfA1a-YFPN152 (HSF-effector), bZIP63-YFPN152 (b-ZIP) or only carrier DNAs (DNA 1: bovine, DNA 2: herring sperm) GFP fluorescence was quantified by flow cytometry. Data were normalized for transformation efficiency using co-transformation of luciferase (LUC) expression plasmid. HS: heat stress treatment (3 hrs, 37°C); RT: room temperature (3Hrs 25°C). The bZIP63-YFPN152 dummy effector was used as a negative control for determining endogenous HSF-activities of protoplasts, which results in similar background levels of the GFP reporter activities as the samples treated only with carrier DNAs, with the dummy effector YFPN152, or without any effector DNA. Heat stress causes a minor increase in GFP reporter activities of theses controls, probably reflecting the promoter activation by the endogenous Hsfs. The HsfA1a-YFPN152 effector shows strongly increased promoter activation with and without heat stress.
6