Gene Expression Profile of THP-1 Cells Treated with Heat-Killed Candida Albicans
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Functional Characterization of the New 8Q21 Asthma Risk Locus
Functional characterization of the new 8q21 Asthma risk locus Cristina M T Vicente B.Sc, M.Sc A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2017 Faculty of Medicine Abstract Genome wide association studies (GWAS) provide a powerful tool to identify genetic variants associated with asthma risk. However, the target genes for many allergy risk variants discovered to date are unknown. In a recent GWAS, Ferreira et al. identified a new association between asthma risk and common variants located on chromosome 8q21. The overarching aim of this thesis was to elucidate the biological mechanisms underlying this association. Specifically, the goals of this study were to identify the gene(s) underlying the observed association and to study their contribution to asthma pathophysiology. Using genetic data from the 1000 Genomes Project, we first identified 118 variants in linkage disequilibrium (LD; r2>0.6) with the sentinel allergy risk SNP (rs7009110) on chromosome 8q21. Of these, 35 were found to overlap one of four Putative Regulatory Elements (PREs) identified in this region in a lymphoblastoid cell line (LCL), based on epigenetic marks measured by the ENCODE project. Results from analysis of gene expression data generated for LCLs (n=373) by the Geuvadis consortium indicated that rs7009110 is associated with the expression of only one nearby gene: PAG1 - located 732 kb away. PAG1 encodes a transmembrane adaptor protein localized to lipid rafts, which is highly expressed in immune cells. Results from chromosome conformation capture (3C) experiments showed that PREs in the region of association physically interacted with the promoter of PAG1. -

1 INVITED REVIEW Mechanisms of Gasdermin Family Members in Inflammasome Signaling and Cell Death Shouya Feng,* Daniel Fox,* Si M
INVITED REVIEW Mechanisms of Gasdermin family members in inflammasome signaling and cell death Shouya Feng,* Daniel Fox,* Si Ming Man Department of Immunology and Infectious Disease, The John Curtin School of Medical Research, The Australian National University, Canberra, Australia. * S.F. and D.F. equally contributed to this work Correspondence to Si Ming Man: Department of Immunology and Infectious Disease, The John Curtin School of Medical Research, The Australian National University, Canberra, 2601, Australia. [email protected] 1 Abstract The Gasdermin (GSDM) family consists of Gasdermin A (GSDMA), Gasdermin B (GSDMB), Gasdermin C (GSDMC), Gasdermin D (GSDMD), Gasdermin E (GSDME) and Pejvakin (PJVK). GSDMD is activated by inflammasome-associated inflammatory caspases. Cleavage of GSDMD by human or mouse caspase-1, human caspase-4, human caspase-5, and mouse caspase-11, liberates the N-terminal effector domain from the C-terminal inhibitory domain. The N-terminal domain oligomerizes in the cell membrane and forms a pore of 10-16 nm in diameter, through which substrates of a smaller diameter, such as interleukin (IL)-1β and IL- 18, are secreted. The increasing abundance of membrane pores ultimately leads to membrane rupture and pyroptosis, releasing the entire cellular content. Other than GSDMD, the N-terminal domain of all GSDMs, with the exception of PJVK, have the ability to form pores. There is evidence to suggest that GSDMB and GSDME are cleaved by apoptotic caspases. Here, we review the mechanistic functions of GSDM proteins with respect to their expression and signaling profile in the cell, with more focused discussions on inflammasome activation and cell death. -

Nº Ref Uniprot Proteína Péptidos Identificados Por MS/MS 1 P01024
Document downloaded from http://www.elsevier.es, day 26/09/2021. This copy is for personal use. Any transmission of this document by any media or format is strictly prohibited. Nº Ref Uniprot Proteína Péptidos identificados 1 P01024 CO3_HUMAN Complement C3 OS=Homo sapiens GN=C3 PE=1 SV=2 por 162MS/MS 2 P02751 FINC_HUMAN Fibronectin OS=Homo sapiens GN=FN1 PE=1 SV=4 131 3 P01023 A2MG_HUMAN Alpha-2-macroglobulin OS=Homo sapiens GN=A2M PE=1 SV=3 128 4 P0C0L4 CO4A_HUMAN Complement C4-A OS=Homo sapiens GN=C4A PE=1 SV=1 95 5 P04275 VWF_HUMAN von Willebrand factor OS=Homo sapiens GN=VWF PE=1 SV=4 81 6 P02675 FIBB_HUMAN Fibrinogen beta chain OS=Homo sapiens GN=FGB PE=1 SV=2 78 7 P01031 CO5_HUMAN Complement C5 OS=Homo sapiens GN=C5 PE=1 SV=4 66 8 P02768 ALBU_HUMAN Serum albumin OS=Homo sapiens GN=ALB PE=1 SV=2 66 9 P00450 CERU_HUMAN Ceruloplasmin OS=Homo sapiens GN=CP PE=1 SV=1 64 10 P02671 FIBA_HUMAN Fibrinogen alpha chain OS=Homo sapiens GN=FGA PE=1 SV=2 58 11 P08603 CFAH_HUMAN Complement factor H OS=Homo sapiens GN=CFH PE=1 SV=4 56 12 P02787 TRFE_HUMAN Serotransferrin OS=Homo sapiens GN=TF PE=1 SV=3 54 13 P00747 PLMN_HUMAN Plasminogen OS=Homo sapiens GN=PLG PE=1 SV=2 48 14 P02679 FIBG_HUMAN Fibrinogen gamma chain OS=Homo sapiens GN=FGG PE=1 SV=3 47 15 P01871 IGHM_HUMAN Ig mu chain C region OS=Homo sapiens GN=IGHM PE=1 SV=3 41 16 P04003 C4BPA_HUMAN C4b-binding protein alpha chain OS=Homo sapiens GN=C4BPA PE=1 SV=2 37 17 Q9Y6R7 FCGBP_HUMAN IgGFc-binding protein OS=Homo sapiens GN=FCGBP PE=1 SV=3 30 18 O43866 CD5L_HUMAN CD5 antigen-like OS=Homo -

Novel Signature Genes for Human Left Ventricle Cardiomyopathies Identifed by Weighted Co- Expression Network Analysis (WGCNA)
Novel Signature Genes for Human Left Ventricle Cardiomyopathies Identied by Weighted Co- expression Network Analysis (WGCNA) Jiao Tian Kunming Institute of Botany Chinese Academy of Sciences Zhengyuan Wu Kunming Institute of Botany Chinese Academy of Sciences Yaqi Zhang Kunming Institute of Botany Chinese Academy of Sciences Yingying He Yunnan University SHUBAI LIU ( [email protected] ) Kunming Institute of Botany Chinese Academy of Sciences Research Keywords: Heart failure, Cardiomyopathy, Ventricle cardiomyocytes, WGCNA, Signature gene Posted Date: October 8th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-86136/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/29 Abstract Background Cardiomyopathy, a heart disease that arises from different etiologies, brings a huge healthcare burden to the global society. Identication of biomarkers can be very useful for early diagnosis of cardiomyopathy, interruption of the disease procession to heart failure, and decrement of the mortality. Methods Clinical cases of cardiomyopathy were screened out of independently investigations from the genomic database. Exploration of its whole transcription disorder pattern by WGCNA (Weighted Gene Co- expression Network Analysis) to discover the signature genes for different subtypes of cardiomyopathy. The hub genes and key pathways, which are correlated to cardiomyopathy traits, were identied through co-expression and protein-protein interaction (PPI) networks enrichment analysis. Discovered hub genes were blast through the Cardiovascular Disease Portal to verify functions related to human cardiomyopathies. Results Three common axes of signature genes were discovered for ve subtypes of cardiomyopathy: 1) Four genes (MDM4, CFLAR, RPS6KB1, PKD1L2) were common for ischemic and ischemic cardiomyopathy subgroups; 2) Subtypes of cardiomyopathy (ischemic, post. -

Quantifying the Mechanisms of Domain Gain in Animal Proteins Marija Buljan*, Adam Frankish, Alex Bateman
Buljan et al. Genome Biology 2010, 11:R74 http://genomebiology.com/2010/11/7/R74 RESEARCH Open Access Quantifying the mechanisms of domain gain in animal proteins Marija Buljan*, Adam Frankish, Alex Bateman Abstract Background: Protein domains are protein regions that are shared among different proteins and are frequently functionally and structurally independent from the rest of the protein. Novel domain combinations have a major role in evolutionary innovation. However, the relative contributions of the different molecular mechanisms that underlie domain gains in animals are still unknown. By using animal gene phylogenies we were able to identify a set of high confidence domain gain events and by looking at their coding DNA investigate the causative mechanisms. Results: Here we show that the major mechanism for gains of new domains in metazoan proteins is likely to be gene fusion through joining of exons from adjacent genes, possibly mediated by non-allelic homologous recombination. Retroposition and insertion of exons into ancestral introns through intronic recombination are, in contrast to previous expectations, only minor contributors to domain gains and have accounted for less than 1% and 10% of high confidence domain gain events, respectively. Additionally, exonization of previously non-coding regions appears to be an important mechanism for addition of disordered segments to proteins. We observe that gene duplication has preceded domain gain in at least 80% of the gain events. Conclusions: The interplay of gene duplication and domain gain demonstrates an important mechanism for fast neofunctionalization of genes. Background molecular mechanisms that underlie gains of new Protein domains are fundamental and largely indepen- domains in animals has been more challenging [3]. -

Setd1 Histone 3 Lysine 4 Methyltransferase Complex Components in Epigenetic Regulation
SETD1 HISTONE 3 LYSINE 4 METHYLTRANSFERASE COMPLEX COMPONENTS IN EPIGENETIC REGULATION Patricia A. Pick-Franke Submitted to the faculty of the University Graduate School in partial fulfillment of the requirements for the degree Master of Science in the Department of Biochemistry and Molecular Biology Indiana University December 2010 Accepted by the Faculty of Indiana University, in partial fulfillment of the requirements for the degree of Master of Science. _____________________________________ David Skalnik, Ph.D., Chair _____________________________________ Kristin Chun, Ph.D. Master’s Thesis Committee _____________________________________ Simon Rhodes, Ph.D. ii DEDICATION This thesis is dedicated to my sons, Zachary and Zephaniah who give me great joy, hope and continuous inspiration. I can only hope that I successfully set a good example demonstrating that one can truly accomplish anything, if you never give up and reach for your dreams. iii ACKNOWLEDGEMENTS I would like to thank my committee members Dr. Skalnik, Dr. Chun and Dr. Rhodes for allowing me to complete this dissertation. They have been incredibly generous with their flexibility. I must make a special thank you to Jeanette McClintock, who willingly gave her expertise in statistical analysis with the Cfp1 microarray data along with encouragement, support and guidance to complete this work. I would like to thank Courtney Tate for her ceaseless willingness to share ideas, and her methods and materials, and Erika Dolbrota for her generous instruction as well as the name of a good doctor. I would also like to acknowledge the superb mentorship of Dr. Jeon Heong Lee, PhD and the contagious passion and excitement for the life of science of Dr. -
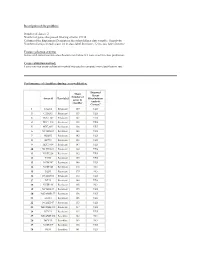
2 Number of Genes That Passed Filtering Criteria
Description of the problem: Number of classes: 2 Number of genes that passed filtering criteria: 25338 Column of the Experiment Descriptors sheet that defines class variable: Sensitivity Number of arrays in each class: 24 in class label Resistant , 32 in class label Sensitive Feature selection criteria: Genes with univariate mis-classfication rate below 0.2 were used for class prediction. Cross-validation method: Leave-one-out cross-validation method was used to compute mis-classification rate. Performance of classifiers during cross-validation. Diagonal Mean Linear Number of Array id Class label Discriminant genes in Analysis classifier Correct? 1 CALU1 Resistant 129 YES 2 CAMA1 Resistant 155 YES 3 HCC1143 Resistant 145 YES 4 HCC1419 Resistant 148 YES 5 HCC2935 Resistant 136 YES 6 NCIH2405 Resistant 148 YES 7 SKBR3 Resistant 142 YES 8 ZR751 Resistant 133 YES 9 HCC1954 Resistant 147 YES 10 NCIH1651 Resistant 139 YES 11 NCIH226 Resistant 142 YES 12 T47D Resistant 139 YES 13 NCIH747 Resistant 146 YES 14 NCIH345 Resistant 216 NO 15 DLD1 Resistant 179 NO 16 NCIH1666 Resistant 154 YES 17 BT20 Resistant 140 YES 18 NCIH196 Resistant 168 NO 19 NCIH1437 Resistant 159 YES 20 MDAMB157 Resistant 158 YES 21 LS123 Resistant 155 YES 22 NCIH2347 Resistant 133 YES 23 MDAMB361 Resistant 137 YES 24 BT474 Resistant 155 YES 25 MDAMB468 Sensitive 182 NO 26 MCF10 Sensitive 169 NO 27 NCIH209 Sensitive 128 YES 28 RKO Sensitive 141 YES 29 HCC70 Sensitive 182 NO 30 LS513 Sensitive 136 YES 31 NCIH1963 Sensitive 122 YES 32 HCC1187 Sensitive 145 YES 33 T84 Sensitive -

Agricultural University of Athens
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΤΩΝ ΖΩΩΝ ΤΜΗΜΑ ΕΠΙΣΤΗΜΗΣ ΖΩΙΚΗΣ ΠΑΡΑΓΩΓΗΣ ΕΡΓΑΣΤΗΡΙΟ ΓΕΝΙΚΗΣ ΚΑΙ ΕΙΔΙΚΗΣ ΖΩΟΤΕΧΝΙΑΣ ΔΙΔΑΚΤΟΡΙΚΗ ΔΙΑΤΡΙΒΗ Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων ΕΙΡΗΝΗ Κ. ΤΑΡΣΑΝΗ ΕΠΙΒΛΕΠΩΝ ΚΑΘΗΓΗΤΗΣ: ΑΝΤΩΝΙΟΣ ΚΟΜΙΝΑΚΗΣ ΑΘΗΝΑ 2020 ΔΙΔΑΚΤΟΡΙΚΗ ΔΙΑΤΡΙΒΗ Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων Genome-wide association analysis and gene network analysis for (re)production traits in commercial broilers ΕΙΡΗΝΗ Κ. ΤΑΡΣΑΝΗ ΕΠΙΒΛΕΠΩΝ ΚΑΘΗΓΗΤΗΣ: ΑΝΤΩΝΙΟΣ ΚΟΜΙΝΑΚΗΣ Τριμελής Επιτροπή: Aντώνιος Κομινάκης (Αν. Καθ. ΓΠΑ) Ανδρέας Κράνης (Eρευν. B, Παν. Εδιμβούργου) Αριάδνη Χάγερ (Επ. Καθ. ΓΠΑ) Επταμελής εξεταστική επιτροπή: Aντώνιος Κομινάκης (Αν. Καθ. ΓΠΑ) Ανδρέας Κράνης (Eρευν. B, Παν. Εδιμβούργου) Αριάδνη Χάγερ (Επ. Καθ. ΓΠΑ) Πηνελόπη Μπεμπέλη (Καθ. ΓΠΑ) Δημήτριος Βλαχάκης (Επ. Καθ. ΓΠΑ) Ευάγγελος Ζωίδης (Επ.Καθ. ΓΠΑ) Γεώργιος Θεοδώρου (Επ.Καθ. ΓΠΑ) 2 Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων Περίληψη Σκοπός της παρούσας διδακτορικής διατριβής ήταν ο εντοπισμός γενετικών δεικτών και υποψηφίων γονιδίων που εμπλέκονται στο γενετικό έλεγχο δύο τυπικών πολυγονιδιακών ιδιοτήτων σε κρεοπαραγωγικά ορνίθια. Μία ιδιότητα σχετίζεται με την ανάπτυξη (σωματικό βάρος στις 35 ημέρες, ΣΒ) και η άλλη με την αναπαραγωγική -

Network-Based Analysis of Key Regulatory Genes Implicated in Type
www.nature.com/scientificreports OPEN Network‑based analysis of key regulatory genes implicated in Type 2 Diabetes Mellitus and Recurrent Miscarriages in Turner Syndrome Anam Farooqui1, Alaa Alhazmi2, Shaful Haque3, Naaila Tamkeen4, Mahboubeh Mehmankhah1, Safa Tazyeen1, Sher Ali5 & Romana Ishrat1* The information on the genotype–phenotype relationship in Turner Syndrome (TS) is inadequate because very few specifc candidate genes are linked to its clinical features. We used the microarray data of TS to identify the key regulatory genes implicated with TS through a network approach. The causative factors of two common co‑morbidities, Type 2 Diabetes Mellitus (T2DM) and Recurrent Miscarriages (RM), in the Turner population, are expected to be diferent from that of the general population. Through microarray analysis, we identifed nine signature genes of T2DM and three signature genes of RM in TS. The power‑law distribution analysis showed that the TS network carries scale‑free hierarchical fractal attributes. Through local‑community‑paradigm (LCP) estimation we fnd that a strong LCP is also maintained which means that networks are dynamic and heterogeneous. We identifed nine key regulators which serve as the backbone of the TS network. Furthermore, we recognized eight interologs functional in seven diferent organisms from lower to higher levels. Overall, these results ofer few key regulators and essential genes that we envisage have potential as therapeutic targets for the TS in the future and the animal models studied here may prove useful in the validation of such targets. Te medical systems and scientists throughout the world are under an unprecedented challenge to meet the medical needs of much of the world’s population that are sufering from chromosomal anomalies. -

Wo2018/232195
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/232195 Al 20 December 2018 (20.12.2018) W !P O PCT (51) International Patent Classification: MANIAN, Ayshwarya; 77 Massachusetts Ave., Cam A61K 31/00 (2006.01) C07K 16/28 (2006.01) bridge, MA 02139 (US). C07K 14/705 (2006.01) C12Q 1/68 (2018.01) (72) Inventor: ROZENBLATT-ROSEN, Orit; 415 Main C07K 16/24 (2006.01) G06F 19/00 (2018.01) Street, Cambridge, MA 02142 (US). (21) International Application Number: (74) Agent: NIX, F., Brent et al.; Johnson, Marcou & Isaacs, PCT/US20 18/037662 LLC, P.O. Box 691, Hoschton, GA 30548 (US). (22) International Filing Date: (81) Designated States (unless otherwise indicated, for every 14 June 2018 (14.06.2018) kind of national protection available): AE, AG, AL, AM, (25) Filing Language: English AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, (26) Publication Langi English DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, (30) Priority Data: HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, 62/5 19,788 14 June 2017 (14.06.2017) US KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, [US/US]; (71) Applicants: THE BROAD INSTITUTE, INC. OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, 415 Main Street, Cambridge, MA 02142 (US). -

Genetic Variation and Functional Analysis of the Cardiomedin Gene
TECHNISCHE UNIVERSITÄT MÜNCHEN LEHRSTUHL FÜR EXPERIMENTELLE GENETIK Genetic Variation and Functional Analysis of the Cardiomedin Gene Zasie Susanne Schäfer Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzende: Univ.-Prof. A. Schnieke, Ph.D. Prüfer der Dissertation: 1. apl. Prof. Dr. J. Adamski 2. Univ.-Prof. Dr. Dr. H.-R. Fries 3. Univ.-Prof. Dr. Th. Meitinger Die Dissertation wurde am. 31.05.2011 bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 02.04.2012 angenommen. Table of Contents Table of contents Abbreviations ........................................................................................................................ 7 1. Summary ..........................................................................................................................10 Zusammenfassung ...............................................................................................................11 2. Introduction ......................................................................................................................12 2.1 Genome-wide association studies (GWAS) and post-GWAS functional genomics ......12 2.2 Genetic influences on cardiac repolarization and sudden cardiac death syndrome in GWAS and the chromosome -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses by a Synthetic TLR3 Ligand Mitigates
Downloaded from http://www.jimmunol.org/ by guest on September 28, 2021 Inducing is online at: average * The Journal of Immunology published online 16 August 2013 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2013/08/16/jimmun ol.1300376 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 28, 2021. Published August 16, 2013, doi:10.4049/jimmunol.1300376 The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN.