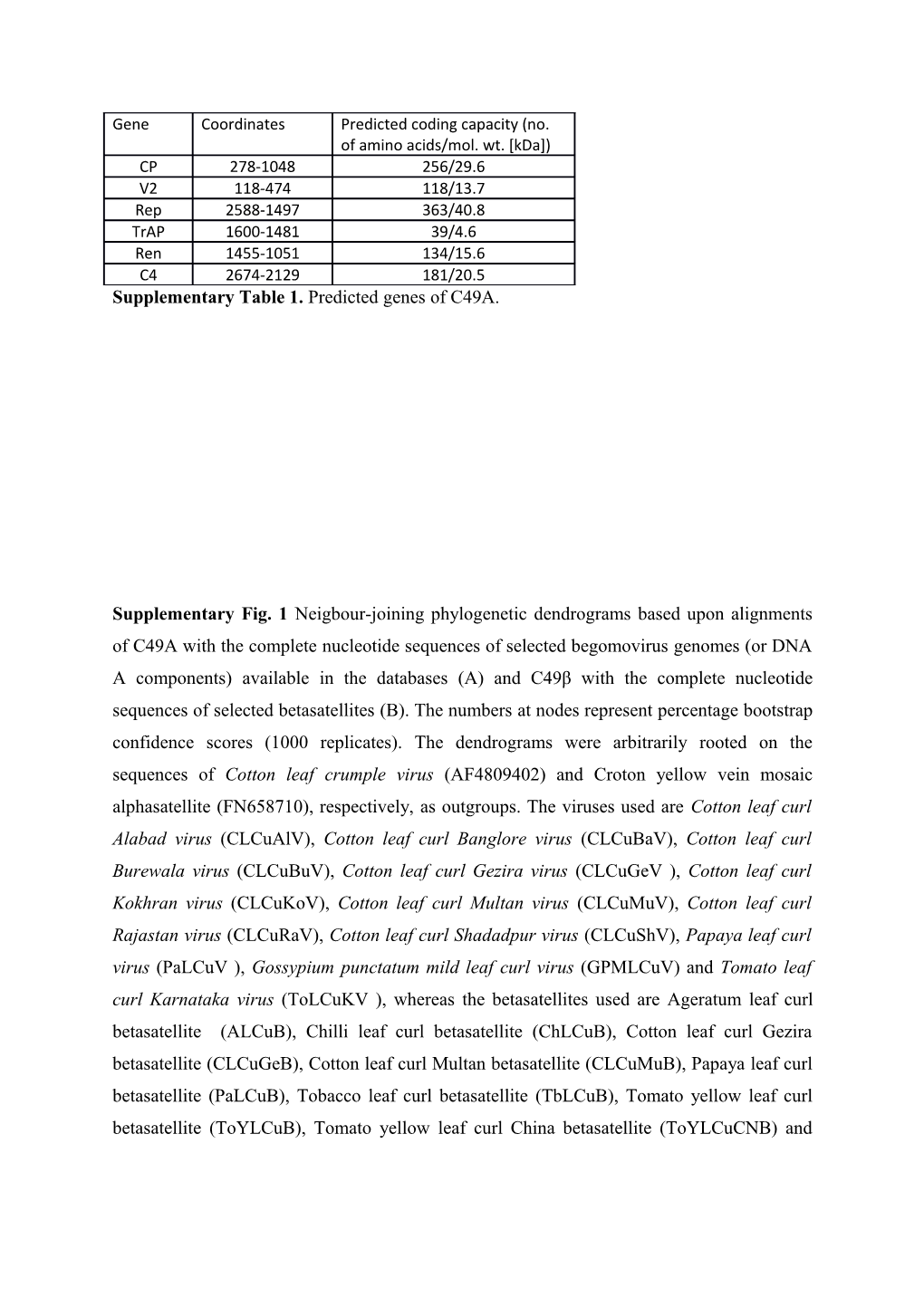
Gene Coordinates Predicted coding capacity (no. of amino acids/mol. wt. [kDa]) CP 278-1048 256/29.6 V2 118-474 118/13.7 Rep 2588-1497 363/40.8 TrAP 1600-1481 39/4.6 Ren 1455-1051 134/15.6 C4 2674-2129 181/20.5 Supplementary Table 1. Predicted genes of C49A.
Supplementary Fig. 1 Neigbour-joining phylogenetic dendrograms based upon alignments of C49A with the complete nucleotide sequences of selected begomovirus genomes (or DNA A components) available in the databases (A) and C49β with the complete nucleotide sequences of selected betasatellites (B). The numbers at nodes represent percentage bootstrap confidence scores (1000 replicates). The dendrograms were arbitrarily rooted on the sequences of Cotton leaf crumple virus (AF4809402) and Croton yellow vein mosaic alphasatellite (FN658710), respectively, as outgroups. The viruses used are Cotton leaf curl Alabad virus (CLCuAlV), Cotton leaf curl Banglore virus (CLCuBaV), Cotton leaf curl Burewala virus (CLCuBuV), Cotton leaf curl Gezira virus (CLCuGeV ), Cotton leaf curl Kokhran virus (CLCuKoV), Cotton leaf curl Multan virus (CLCuMuV), Cotton leaf curl Rajastan virus (CLCuRaV), Cotton leaf curl Shadadpur virus (CLCuShV), Papaya leaf curl virus (PaLCuV ), Gossypium punctatum mild leaf curl virus (GPMLCuV) and Tomato leaf curl Karnataka virus (ToLCuKV ), whereas the betasatellites used are Ageratum leaf curl betasatellite (ALCuB), Chilli leaf curl betasatellite (ChLCuB), Cotton leaf curl Gezira betasatellite (CLCuGeB), Cotton leaf curl Multan betasatellite (CLCuMuB), Papaya leaf curl betasatellite (PaLCuB), Tobacco leaf curl betasatellite (TbLCuB), Tomato yellow leaf curl betasatellite (ToYLCuB), Tomato yellow leaf curl China betasatellite (ToYLCuCNB) and Tomato yellow leaf curl Thailand betasatellite (ToYLCuThB). In each case the database accession number is given.
Supplementary Fig. 2 Sequence of C49A surrounding the overlap of the Rep, TrAP and REn genes highlighting coding sequences, start and stop codons (as indicated by the key at the top). Note the truncated (predicted 39 amino acid coding capacity) TrAP gene.