Identification of Potential Common Cyclic‑Di‑GMP Regulated Biomarkers Among Three Gram Negative Bacteria Via Transcriptomic and Metabolomic Profiling
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Peter D Karp Ingrid Keseler An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Anamika Kothari Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000223375Cyc: Ketogulonicigenium vulgare WSH-001 (GCF_000223375) Cellular Overview Connections between pathways are omitted for legibility. Ron Caspi a sulfonate a sulfonate L-arabinose L-arabinose L-arabinose L-arabinose L-arabinose spermidine nitrate nitrate phosphate D-galactose D-galactose D-galactose D-galactose D-galactose 3+ 3+ putrescine L-ornithine L-ornithine L-ornithine L-ornithine a sulfonate a sulfonate a sulfonate a sulfonate a sulfonate hydrogencarbonate hydrogencarbonate a sulfonate Fe Fe an amino glycine betaine D-allose D-allose D-allose D-allose D-allose spermidine spermidine spermidine sn-glycerol sn-glycerol spermidine lys lys lys lys molybdate molybdate molybdate molybdate nitrate nitrate nitrate nitrate nitrate nitrate thiosulfate thiosulfate thiosulfate thiosulfate thiosulfate thiosulfate spermidine spermidine ammonium a glycerophosphodiester acid L-carnitine phosphate phosphate phosphate benzoate hydrogencarbonate hydrogencarbonate hydrogencarbonate hydrogencarbonate hydrogencarbonate hydrogencarbonate -

Handbook of Essential Oils: Science, Technology, and Applications
Handbook of ESSENTIAL Science, Technology, OILS and Applications Handbook of ESSENTIAL Science, Technology, OILS and Applications Edited by K. Hüsnü Can Bas¸er Gerhard Buchbauer Boca Raton London New York CRC Press is an imprint of the Taylor & Francis Group, an informa business CRC Press Taylor & Francis Group 6000 Broken Sound Parkway NW, Suite 300 Boca Raton, FL 33487-2742 © 2010 by Taylor and Francis Group, LLC CRC Press is an imprint of Taylor & Francis Group, an Informa business No claim to original U.S. Government works Printed in the United States of America on acid-free paper 10 9 8 7 6 5 4 3 2 1 International Standard Book Number: 978-1-4200-6315-8 (Hardback) This book contains information obtained from authentic and highly regarded sources. Reasonable efforts have been made to publish reliable data and information, but the author and publisher cannot assume responsibility for the valid- ity of all materials or the consequences of their use. The authors and publishers have attempted to trace the copyright holders of all material reproduced in this publication and apologize to copyright holders if permission to publish in this form has not been obtained. If any copyright material has not been acknowledged please write and let us know so we may rectify in any future reprint. Except as permitted under U.S. Copyright Law, no part of this book may be reprinted, reproduced, transmitted, or uti- lized in any form by any electronic, mechanical, or other means, now known or hereafter invented, including photocopy- ing, microfilming, and recording, or in any information storage or retrieval system, without written permission from the publishers. -

Novel Approaches for Using Dehydrogenases and Ene-Reductases for Organic Synthesis
Novel approaches for using dehydrogenases and ene-reductases for organic synthesis Serena GARGIULO Novel approaches for using dehydrogenases and ene-reductases for organic synthesis Proefschrift ter verkrijging van de graad van doctor aan de Technische Universiteit Delft, op gezag van de Rector Magnificus prof. ir. K.C.A.M. Luyben, voorzitter van het College voor Promoties, in het openbaar te verdedigen op dinsdag 24 februari 2015 om 10.00 uur door Serena GARGIULO Master of Science in Molecular and Industrial Biotechnology van Università degli Studi di Napoli “Federico II”, Italië, geboren te Napels, Italië Dit proefschrift is goedgekeurd door de promotor: Prof Dr. I.W.C.E. Arends Copromotor Dr.F.Hollmann Samenstelling promotiecommissie: Rector Magnificus, voorzitter Prof Dr. I. W. C. E. Arends Technische Universiteit Delft, promotor Dr. F. Hollmann Technische Universiteit Delft, supervisor Prof. Dr. S. De Vries Technische Universiteit Delft Prof. Dr. R. Piccoli Università degli Studi di Napoli “Federico II” Prof. Dr. W. van Berkel Wageningen Universiteit Prof. Dr. G. Grogan University of York Dr. Stephan Luetz Novartis Pharma Prof. Dr. E. J. R. Sudholter Technische Universiteit Delft, reservelid ISBN 978-94-6108-919-9 The research reported in this thesis was supported by the Marie Curie Initial Training Network BIOTRAINS, financed by the European Union through the 7th Framework People Programme (grant agreement number 238531) Ai miei Angeli, in terra e in cielo, a chi ha creduto in me Table of contents Chapter 1 Introduction……………………………………………………………………………1 Chapter 2 A photoenzymatic system for alcohol oxidation……………………………………..29 Chapter 3 A biocatalytic redox isomerization…………………………………………………...47 Chapter 4 Synthetic nicotinamide cofactors for biocatalytic reduction of activated C=C bonds…………………………………………………………………………...67 Chapter 5 Structure of the alcohol dehydrogenase from Thermus sp . -

Microbial Monoterpene Transformations—A Review
REVIEW ARTICLE published: 15 July 2014 doi: 10.3389/fmicb.2014.00346 Microbial monoterpene transformations—a review Robert Marmulla and Jens Harder* Department of Microbiology, Max Planck Institute for Marine Microbiology, Bremen, Germany Edited by: Isoprene and monoterpenes constitute a significant fraction of new plant biomass. Colin Murrell, University of East Emission rates into the atmosphere alone are estimated to be over 500 Tg per year. These Anglia, UK natural hydrocarbons are mineralized annually in similar quantities. In the atmosphere, Reviewed by: abiotic photochemical processes cause lifetimes of minutes to hours. Microorganisms Terry John McGenity, University of Essex, UK encounter isoprene, monoterpenes, and other volatiles of plant origin while living in and Andrew Crombie, University of East on plants, in the soil and in aquatic habitats. Below toxic concentrations, the compounds Anglia, UK can serve as carbon and energy source for aerobic and anaerobic microorganisms. *Correspondence: Besides these catabolic reactions, transformations may occur as part of detoxification Jens Harder, Max Planck Institute processes. Initial transformations of monoterpenes involve the introduction of functional for Marine Microbiology, Celsiusstr. 1, Bremen 28359, Germany groups, oxidation reactions, and molecular rearrangements catalyzed by various enzymes. e-mail: [email protected] Pseudomonas and Rhodococcus strains and members of the genera Castellaniella and Thauera have become model organisms for the elucidation of biochemical pathways. We review here the enzymes and their genes together with microorganisms known for a monoterpene metabolism, with a strong focus on microorganisms that are taxonomically validly described and currently available from culture collections. Metagenomes of microbiomes with a monoterpene-rich diet confirmed the ecological relevance of monoterpene metabolism and raised concerns on the quality of our insights based on the limited biochemical knowledge. -

All Enzymes in BRENDA™ the Comprehensive Enzyme Information System
All enzymes in BRENDA™ The Comprehensive Enzyme Information System http://www.brenda-enzymes.org/index.php4?page=information/all_enzymes.php4 1.1.1.1 alcohol dehydrogenase 1.1.1.B1 D-arabitol-phosphate dehydrogenase 1.1.1.2 alcohol dehydrogenase (NADP+) 1.1.1.B3 (S)-specific secondary alcohol dehydrogenase 1.1.1.3 homoserine dehydrogenase 1.1.1.B4 (R)-specific secondary alcohol dehydrogenase 1.1.1.4 (R,R)-butanediol dehydrogenase 1.1.1.5 acetoin dehydrogenase 1.1.1.B5 NADP-retinol dehydrogenase 1.1.1.6 glycerol dehydrogenase 1.1.1.7 propanediol-phosphate dehydrogenase 1.1.1.8 glycerol-3-phosphate dehydrogenase (NAD+) 1.1.1.9 D-xylulose reductase 1.1.1.10 L-xylulose reductase 1.1.1.11 D-arabinitol 4-dehydrogenase 1.1.1.12 L-arabinitol 4-dehydrogenase 1.1.1.13 L-arabinitol 2-dehydrogenase 1.1.1.14 L-iditol 2-dehydrogenase 1.1.1.15 D-iditol 2-dehydrogenase 1.1.1.16 galactitol 2-dehydrogenase 1.1.1.17 mannitol-1-phosphate 5-dehydrogenase 1.1.1.18 inositol 2-dehydrogenase 1.1.1.19 glucuronate reductase 1.1.1.20 glucuronolactone reductase 1.1.1.21 aldehyde reductase 1.1.1.22 UDP-glucose 6-dehydrogenase 1.1.1.23 histidinol dehydrogenase 1.1.1.24 quinate dehydrogenase 1.1.1.25 shikimate dehydrogenase 1.1.1.26 glyoxylate reductase 1.1.1.27 L-lactate dehydrogenase 1.1.1.28 D-lactate dehydrogenase 1.1.1.29 glycerate dehydrogenase 1.1.1.30 3-hydroxybutyrate dehydrogenase 1.1.1.31 3-hydroxyisobutyrate dehydrogenase 1.1.1.32 mevaldate reductase 1.1.1.33 mevaldate reductase (NADPH) 1.1.1.34 hydroxymethylglutaryl-CoA reductase (NADPH) 1.1.1.35 3-hydroxyacyl-CoA -

(12) United States Patent (10) Patent No.: US 9,017,983 B2 Burgard Et Al
US009017983B2 (12) United States Patent (10) Patent No.: US 9,017,983 B2 Burgard et al. (45) Date of Patent: Apr. 28, 2015 (54) ORGANISMS FOR THE PRODUCTION OF 5,521,075 A 5/1996 Guettler et al. 13-BUTANEDOL 5,573.931 A 11/1996 Guettler et al. 9 5,616,496 A 4/1997 Frost et al. 5,661,026 A 8/1997 Peoples et al. (75) Inventors: Anthony P. Burgard, Bellefonte, PA 5,686,276 A 1 1/1997 Lafend et al. (US); Mark J. Burk, San Diego, CA 5,700.934. A 12/1997 Wolters et al. (US); Robin E. Osterhout, San Diego, 5,770.435 A 6/1998 Donnelly et al. t sS); Priti Pharkya, San Diego, CA (Continued) FOREIGN PATENT DOCUMENTS (73) Assignee: Genomatica, Inc., San Diego, CA (US) CN 1358 841 T 2002 (*) Notice: Subject to any disclaimer, the term of this EP O 494 O78 7, 1992 patent is extended or adjusted under 35 E. s 29: U.S.C. 154(b) by 654 days. GB 1230276 4f1971 GB 1314126 4f1973 (21) Appl. No.: 12/772,114 GB 1344557 1, 1974 GB 1512751 6, 1978 (22) Filed: Apr. 30, 2010 JP 50 OO6776 1, 1975 WO WO 82,03854 11, 1982 (65) Prior Publication Data (Continued) US 2010/033.0635A1 Dec. 30, 2010 OTHER PUBLICATIONS Related U.S. Application Data Abadjieva et al., “The Yeast ARG7 Gene Product is Autoproteolyzed (60) Provisional application No. 61/174,473, filed on Apr. to Two Subunit Peptides, Yielding Active Ornithine 30, 2009 Acetyltransferase.” J. Biol. -

Is It a European Car Or a Japanese Car? an ERP Study of Diagnostic Information Use in Visual Expertise
Wright State University CORE Scholar Psychology Faculty Publications Psychology 11-2007 Is it a European car or a Japanese car? An ERP Study of Diagnostic Information Use in Visual Expertise Assaf Harel Wright State University - Main Campus, [email protected] Shlomo Bentin Follow this and additional works at: https://corescholar.libraries.wright.edu/psychology Part of the Cognition and Perception Commons, and the Cognitive Psychology Commons Repository Citation Harel, A., & Bentin, S. (2007). Is it a European car or a Japanese car? An ERP Study of Diagnostic Information Use in Visual Expertise. Neural Plasticity, 2007, 30585, 52. https://corescholar.libraries.wright.edu/psychology/254 This Abstract is brought to you for free and open access by the Psychology at CORE Scholar. It has been accepted for inclusion in Psychology Faculty Publications by an authorized administrator of CORE Scholar. For more information, please contact [email protected]. Hindawi Publishing Corporation Neural Plasticity Volume 2007, Article ID 30585, 168 pages doi:10.1155/2007/30585 Meeting Abstracts Abstracts of the 16th Annual Meeting of The Israel Society for Neuroscience Eilat, Israel, November 25–27, 2007 Received 9 October 2007; Accepted 9 October 2007 The Israel Society for Neuroscience—ISFN—was founded in 1993 by a group of Israeli leading scientists conducting research in the area of neurobiology. The primary goal of the society was to promote and disseminate the knowledge and understanding acquired by its members, and to strengthen interactions between them. Since then, the society holds its annual meeting every year in Eilat usually during December. At this annual meetings, the senior Israeli neurobiologists, their teams, and their graduate students, as well as foreign scientists and students, present their recent research findings in platform and poster presentations, and the program of the meeting is mainly based on the 338 received abstracts which are published in this volume. -

Bioenergetics of Mycobacterium: an Emerging Landscape for Drug Discovery
pathogens Review Bioenergetics of Mycobacterium: An Emerging Landscape for Drug Discovery Iram Khan Iqbal †, Sapna Bajeli †, Ajit Kumar Akela and Ashwani Kumar * ID Council of Scientific and Industrial Research, Institute of Microbial Technology, Chandigarh 160036, India; [email protected] (I.K.I.); [email protected] (S.B.); [email protected] (A.K.A.) * Correspondence: [email protected] † These authors contributed equally to this work. Received: 11 January 2018; Accepted: 31 January 2018; Published: 23 February 2018 Abstract: Mycobacterium tuberculosis (Mtb) exhibits remarkable metabolic flexibility that enables it to survive a plethora of host environments during its life cycle. With the advent of bedaquiline for treatment of multidrug-resistant tuberculosis, oxidative phosphorylation has been validated as an important target and a vulnerable component of mycobacterial metabolism. Exploiting the dependence of Mtb on oxidative phosphorylation for energy production, several components of this pathway have been targeted for the development of new antimycobacterial agents. This includes targeting NADH dehydrogenase by phenothiazine derivatives, menaquinone biosynthesis by DG70 and other compounds, terminal oxidase by imidazopyridine amides and ATP synthase by diarylquinolines. Importantly, oxidative phosphorylation also plays a critical role in the survival of persisters. Thus, inhibitors of oxidative phosphorylation can synergize with frontline TB drugs to shorten the course of treatment. In this review, we discuss the oxidative phosphorylation pathway and development of its inhibitors in detail. Keywords: Mycobacterium tuberculosis; bioenergetics; oxidative phosphorylation; antimycobacterials; drugs; bedaquiline; Q203; SQ109; electron transport chain 1. Introduction Tuberculosis (TB) remains a leading cause of death worldwide, with an estimated 1.3 million mortalities in 2016. -

(12) United States Patent (10) Patent No.: US 8,124.387 B2 (51) Int. Cl.
USOO8124387B2 (12) United States Patent (10) Patent No.: US 8,124.387 B2 Stuermer et al. (45) Date of Patent: Feb. 28, 2012 (54) PROCESS FOR THE PRODUCTION OF FOREIGN PATENT DOCUMENTS CTRONELLAL EP 0000315 A1 1, 1979 GB 1476818 6, 1977 (75) Inventors: Rainer Stuermer, Roedersheim-Gronau OTHER PUBLICATIONS (DE); Thomas Friedrich, Darmstadt (DE); Andre Mueller, Vienna (AU): hastiller, A.,et et and al., “Enzymatic J TR Reduction of thes:2. O.B. Unsaturate d Bernhard Hauer, Fussgoenheim (DE): Carbon Bond in Citral”, Journal of Molecular Catalyst B: Enzymatic, Bettina Rosche, Randwick (AU) 2006, vol. 38, pp. 126-130. Williams, R. E., et al., “New Uses for an Old Enzyme'. The Old (73) Assignee: BASFSE, Ludwigshafen (DE) Yellow Enzyme Family of Flavoenzymes'. Microbiology, 2002, vol. 148, pp. 1607-1614. (*) Notice: Subject to any disclaimer, the term of this Vaz, A. D.N. et al., “Old Yellow Enzyme: Aromatization of Cyclic atent is extended or adiusted under 35 Enones and the Mechanism of a Novel Dismutation Reaction'. Bio p chemistry, 1995, vol. 34, pp. 4246-4256. U.S.C. 154(b) by 958 days. Kitzing, K., et al., “The 13 A Crystal Structure of the Flavoprotein YoM reveals a Novel Class of Old Yellow Enzymes'. The Journal of (21) Appl. No.: 12/093,796 Biological Chemistry, 2005, vol. 280, No. 30, 27904-27913. Stott, K., et al., “Old Yellow Enzyme'. The Journal of Biological (22) PCT Filed: Nov. 10, 2006 Chemistry, 1993, vol. 268, No. 9, pp. 6097-6106. Seo, J., et al., “The genome sequence of the ethanologenic bacterium (86). -

(19) United States (12) Patent Application Publication (10) Pub
US 20130244920A1 (19) United States (12) Patent Application Publication (10) Pub. N0.: US 2013/0244920 A1 Lee et al. (43) Pub. Date: Sep. 19, 2013 (54) WATER SOLUBLE COMPOSITIONS (52) US. Cl. INCORPORATING ENZYMES, AND METHOD USPC ......................................... .. 510/392; 264/299 OF MAKING SAME (57) ABSTRACT (76) Inventors: David M. Lee, CroWn Point, IN (US); Jennifer L‘ Sims’ Lowell’ IN (Us) Disclosed herein are Water soluble compositions, such as ?lms, including a mixture of a ?rst Water-soluble resin, an (21) Appl' NO': 13/422’709 enzyme, and an enzyme stabilizer Which comprises a func (22) Filed: Man 16, 2012 tional substrate for the enzyme, methods of making such compositions, and methods of using such compositions, e.g. Publication Classi?cation to make packets containing functional ingredients. The enzymes can include proteases and mixtures of proteases (51) Int. Cl. With other enzymes, and the compositions provide good C11D 3/386 (2006.01) retention of enzyme function following ?lm processing and B29C 39/02 (2006.01) storage. US 2013/0244920 A1 Sep. 19,2013 WATER SOLUBLE COMPOSITIONS preheated to a temperature less than 77° C., optionally in a INCORPORATING ENZYMES, AND METHOD range ofabout 66° C. to about 77° C., or about 74° C.; drying OF MAKING SAME the Water from the cast mixture over a period of less than 24 hours, optionally less than 12 hours, optionally less than 8 FIELD OF THE DISCLOSURE hours, optionally less than 2 hours, optionally less than 1 [0001] The present disclosure relates generally to Water hour, optionally less than 45 minutes, optionally less than 30 soluble ?lms. -
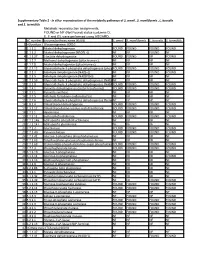
Supplementary Table 2 - in Silico Reconstruction of the Metabolic Pathways of S
Supplementary Table 2 - In silico reconstruction of the metabolic pathways of S. amnii , S. moniliformis , L. buccalis and S. termiditis Metabolic reconstruction assignments, FOUND or NF (Not Found) status (columns D, E, F and G), were performed using ASGARD, EC number Enzyme/pathway name (KEGG) S. amnii S. moniliformis L. buccalis S. termiditis 1 >Glycolysis / Gluconeogenesis 00010 2 1.1.1.1 Alcohol dehydrogenase. FOUND FOUND FOUND FOUND 3 1.1.1.2 Alcohol dehydrogenase (NADP(+)). NF NF FOUND NF 4 1.1.1.27 L-lactate dehydrogenase. FOUND FOUND NF FOUND 5 1.1.2.7 Methanol dehydrogenase (cytochrome c). NF NF NF NF 6 1.1.2.8 Alcohol dehydrogenase (cytochrome c). NF NF NF NF 7 1.2.1.12 Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating).FOUND FOUND FOUND FOUND 8 1.2.1.3 Aldehyde dehydrogenase (NAD(+)). NF NF FOUND FOUND 9 1.2.1.5 Aldehyde dehydrogenase (NAD(P)(+)). NF NF NF NF 10 1.2.1.59 Glyceraldehyde-3-phosphate dehydrogenase (NAD(P)(+))NF (phosphorylating).NF NF NF 11 1.2.1.9 Glyceraldehyde-3-phosphate dehydrogenase (NADP(+)).FOUND FOUND FOUND FOUND 12 1.2.4.1 Pyruvate dehydrogenase (acetyl-transferring). FOUND FOUND FOUND FOUND 13 1.2.7.1 Pyruvate synthase. NF NF NF NF 14 1.2.7.5 Aldehyde ferredoxin oxidoreductase. NF NF NF NF 15 1.2.7.6 Glyceraldehyde-3-phosphate dehydrogenase (ferredoxin).NF NF NF NF 16 1.8.1.4 Dihydrolipoyl dehydrogenase. FOUND FOUND FOUND FOUND 17 2.3.1.12 Dihydrolipoyllysine-residue acetyltransferase. FOUND FOUND FOUND FOUND 18 2.7.1.1 Hexokinase. -

Genetic and Biochemical Characterization of a Novel Monoterpene Ε-Lactone Hydrolase from Rhodococcus Erythropolis DCL14
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Feb. 2001, p. 733–741 Vol. 67, No. 2 0099-2240/01/$04.00ϩ0 DOI: 10.1128/AEM.67.2.733–741.2001 Copyright © 2001, American Society for Microbiology. All Rights Reserved. Genetic and Biochemical Characterization of a Novel Monoterpene ε-Lactone Hydrolase from Rhodococcus erythropolis DCL14 1 1,2 CE´CILE J. B VAN DER VLUGT-BERGMANS AND MARIE¨TJ.VAN DER WERF * Division of Industrial Microbiology, Department of Food Technology and Nutritional Sciences, Wageningen University, Wageningen,1 and Department of Applied Microbiology and Gene Technology, TNO Nutrition and Food Research, Zeist,2 The Netherlands Received 22 August 2000/Accepted 30 November 2000 A monoterpene -lactone hydrolase (MLH) from Rhodococcus erythropolis DCL14, catalyzing the ring open- Downloaded from ing of lactones which are formed during degradation of several monocyclic monoterpenes, including carvone and menthol, was purified to apparent homogeneity. It is a monomeric enzyme of 31 kDa that is active with (4R)-4-isopropenyl-7-methyl-2-oxo-oxepanone and (6R)-6-isopropenyl-3-methyl-2-oxo-oxepanone, lactones de- rived from (4R)-dihydrocarvone, and 7-isopropyl-4-methyl-2-oxo-oxepanone, the lactone derived from men- thone. Both enantiomers of 4-, 5-, 6-, and 7-methyl-2-oxo-oxepanone were converted at equal rates, suggesting that the enzyme is not stereoselective. Maximal enzyme activity was measured at pH 9.5 and 30°C. Determi- nation of the N-terminal amino acid sequence of purified MLH enabled cloning of the corresponding gene by a combination of PCR and colony screening. The gene, designated mlhB (monoterpene lactone hydrolysis), showed up to 43% similarity to members of the GDXG family of lipolytic enzymes.