Activity and Community Composition of Denitrifying Bacteria in Poly(3-Hydroxybutyrate-Co-3-Hydroxyvalerate)-Using Solid-Phase Denitrification Processes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Insight Into the Resistome and Quorum Sensing System of a Divergent Acinetobacter Pittii Isolate from 1 an Untouched Site Of
bioRxiv preprint doi: https://doi.org/10.1101/745182; this version posted November 27, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Insight into the resistome and quorum sensing system of a divergent Acinetobacter pittii isolate from 2 an untouched site of the Lechuguilla Cave 3 4 Han Ming Gan1,2,3*, Peter Wengert4 , Hazel A. Barton5, André O. Hudson4 and Michael A. Savka4 5 1 Centre for Integrative Ecology, School of Life and Environmental Sciences, Deakin University, Geelong 6 3220 ,Victoria, Australia 7 2 Deakin Genomics Centre, Deakin University, Geelong 3220 ,Victoria, Australia 8 3 School of Science, Monash University Malaysia, Bandar Sunway, 47500 Petaling Jaya, Selangor, 9 Malaysia 10 4 Thomas H. Gosnell School of Life Sciences, Rochester Institute of Technology, Rochester, NY, USA 11 5 Department of Biology, University of Akron, Akron, Ohio, USA 12 *Corresponding author 13 Name: Han Ming Gan 14 Email: [email protected] 15 Key words 16 Acinetobacter, quorum sensing, antibiotic resistance 17 18 Abstract 19 Acinetobacter are Gram-negative bacteria belonging to the sub-phyla Gammaproteobacteria, commonly 20 associated with soils, animal feeds and water. Some members of the Acinetobacter have been 21 implicated in hospital-acquired infections, with broad-spectrum antibiotic resistance. Here we report the 22 whole genome sequence of LC510, an Acinetobacter species isolated from deep within a pristine 23 location of the Lechuguilla Cave. -

Denitrification Process Enhancement and Diversity of the Denitrifying
energies Article Denitrification Process Enhancement and Diversity of the Denitrifying Community in the Full Scale Activated Sludge System after Adaptation to Fusel Oil Przemysław Kowal 1 , Sławomir Ciesielski 2,* , Jeremiah Otieno 1, Joanna Barbara Majtacz 1, Krzysztof Czerwionka 1 and Jacek M ˛akinia 1 1 Faculty of Civil and Environmental Engineering, Gdansk University of Technology, Narutowicza 11/12, 80-233 Gdansk, Poland; [email protected] (P.K.); [email protected] (J.O.); [email protected] (J.B.M.); [email protected] (K.C.); [email protected] (J.M.) 2 Department of Environmental Biotechnology, Faculty of Geoengineering, University of Warmia and Mazury in Olsztyn, ul. Słoneczna 45G, 10-709 Olsztyn, Poland * Correspondence: [email protected] Abstract: Implementation of anaerobic digestion of primary sludge in modern wastewater treat- ment plants (WWTPs) limits the availability of organic carbon for denitrification in conventional nitrification-denitrification (N/DN) systems. In order to ensure efficient denitrification, dosage of the external carbon source is commonly undertaken. However, application of commercial products, such us ethanol or acetate, greatly increases operational costs. As such, inexpensive and efficient alternative carbon sources are strongly desirable. In this study, the use of the fusel oil, a by-product from the distillery industry, was validated in terms of the denitrification process enhancement and Citation: Kowal, P.; Ciesielski, S.; impact on the activated sludge bacterial community structure. The experiment was conducted at a Otieno, J.; Majtacz, J.B.; Czerwionka, full scale biological nutrient removal facility (210,000 PE), in the set of the two technological lines: the K.; M ˛akinia,J. -

Microbial Community Response to Heavy and Light Crude Oil in the Great Lakes
Microbial Community Response to Heavy and Light Crude Oil in the Great Lakes Stephen Techtmann 10/24/19 Microbial Sensors Techtmann Lab @ MTU Investigating the applications of environmental microbial communities Hydraulic Fracturing Related Antibiotic Resistance Oil Bioremediation Techtmann Lab @ MTU Overview • Background on oil biodegradation • Microbial response to light and heavy crude oil in the Great Lakes • Machine learning for prediction of contamination in the Great Lakes. Oil Spills Deepwater Horizon Enbridge Line 6B Deepwater Horizon Oil Spill • 4,1000,000 bbl of oil released • Light Sweet Crude oil released • April 20, 2010 • 1101.7 miles of shoreline oiled Atlas and Hazen 2011 Enbridge Line 6B Spill – Marshall MI • 20,082 bbl of oil released • Diluted Bitumen • July 26, 2010 • 70 miles of shoreline oiled https://www.mlive.com/news/kalamazoo/2010/07/state_of_emergency_declared_as.html Oil Transmissions Pipelines in the Great Lakes Region Line 5: • 645 miles from Superior WI to Sarnia Ontario • 540,000 barrels per day • Light crude and natural gas liquids (NGLs) Crude oil Oil types and API Gravity Microbes and Biotechnology (Bioremediation) Low cost input Microbe High value output Decreased Cost Contaminant Increased Efficiency Carbon dioxide or non- toxic daughter products Carbon dioxide Microbial Biomass Petroleum Microbe Daughter Products Water Microbial Ecology and Biotechnology Low cost input Microbe High value output Decreased Cost/Increased Efficiency Complex input Input A Microbe Microbe Output A Input B Microbe Output -

In Situ Electrochemical Studies of the Terrestrial Deep Subsurface Biosphere at the Sanford
bioRxiv preprint doi: https://doi.org/10.1101/555474; this version posted February 20, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 In Situ Electrochemical Studies of the Terrestrial Deep Subsurface Biosphere at the Sanford 2 Underground Research Facility, South Dakota, USA 3 4 Yamini Jangir,a Amruta A. Karbelkar,b Nicole M. Beedle,c Laura A. Zinke,d Greg Wanger,d 5 Cynthia M. Anderson,e Brandi Kiel Reese,f Jan P. Amend,c,d and Mohamed Y. El-Naggar,a,b,c#, 6 7 Department of Physics and Astronomy, University of Southern California, Los Angeles, 8 California, USAa; 9 Department of Chemistry, University of Southern California, Los Angeles, California, USAb; 10 Department of Biological Sciences, University of Southern California, Los Angeles, California, 11 USAc; 12 Department of Earth Science, University of Southern California, Los Angeles, California, USAd; 13 Center for the Conservation of Biological Resources, Black Hills State University, Spearfish, 14 South Dakota, USAe 15 Department of Life Sciences, Texas A&M University, Corpus Christi, Texas, USAf; 16 17 Running Head: [limit: 54 characters and spaces] 18 19 #Address correspondence to Mohamed Y. El-Naggar, [email protected]. 20 21 22 1 bioRxiv preprint doi: https://doi.org/10.1101/555474; this version posted February 20, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -
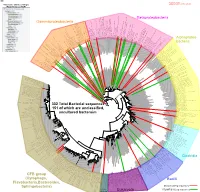
Bacteria Clostridia Bacilli Eukaryota CFB Group
AM935842.1.1361 uncultured Burkholderiales bacterium Class Betaproteobacteria AY283260.1.1552 Alcaligenes sp. PCNB−2 Class Betaproteobacteria AM934953.1.1374 uncultured Burkholderiales bacterium Class Betaproteobacteria AJ581593.1.1460 uncultured betaAM936569.1.1351 proteobacterium uncultured Class Betaproteobacteria Derxia sp. Class Betaproteobacteria AJ581621.1.1418 uncultured beta proteobacterium Class Betaproteobacteria DQ248272.1.1498 uncultured soil bacterium soil uncultured DQ248272.1.1498 DQ248235.1.1498 uncultured soil bacterium RS49 DQ248270.1.1496 uncultured soil bacterium DQ256489.1.1211 Variovorax paradoxus Class Betaproteobacteria Class paradoxus Variovorax DQ256489.1.1211 AF523053.1.1486 uncultured Comamonadaceae bacterium Class Betaproteobacteria AY706442.1.1396 uncultured bacterium uncultured AY706442.1.1396 AJ536763.1.1422 uncultured bacterium CS000359.1.1530 Variovorax paradoxus Class Betaproteobacteria Class paradoxus Variovorax CS000359.1.1530 AY168733.1.1411 uncultured bacterium AJ009470.1.1526 uncultured bacterium SJA−62 Class Betaproteobacteria Class SJA−62 bacterium uncultured AJ009470.1.1526 AY212561.1.1433 uncultured bacterium D16212.1.1457 Rhodoferax fermentans Class Betaproteobacteria Class fermentans Rhodoferax D16212.1.1457 AY957894.1.1546 uncultured bacterium AJ581620.1.1452 uncultured beta proteobacterium Class Betaproteobacteria RS76 AY625146.1.1498 uncultured bacterium RS65 DQ316832.1.1269 uncultured beta proteobacterium Class Betaproteobacteria DQ404909.1.1513 uncultured bacterium uncultured DQ404909.1.1513 AB021341.1.1466 bacterium rM6 AJ487020.1.1500 uncultured bacterium uncultured AJ487020.1.1500 RS7 RS86RC AF364862.1.1425 bacterium BA128 Class Betaproteobacteria AY957931.1.1529 uncultured bacterium uncultured AY957931.1.1529 CP000884.723807.725332 Delftia acidovorans SPH−1 Class Betaproteobacteria AY957923.1.1520 uncultured bacterium uncultured AY957923.1.1520 RS18 AY957918.1.1527 uncultured bacterium uncultured AY957918.1.1527 AY945883.1.1500 uncultured bacterium AF526940.1.1489 uncultured Ralstonia sp. -

Methylene Blue Decolorizing Bacteria Isolated from Water Sewage in Yogyakarta, Indonesia
BIODIVERSITAS ISSN: 1412-033X Volume 21, Number 3, March 2020 E-ISSN: 2085-4722 Pages: 1136-1141 DOI: 10.13057/biodiv/d210338 Methylene blue decolorizing bacteria isolated from water sewage in Yogyakarta, Indonesia MICHELLE, RACHEL ARVY NABASA SIREGAR, ASTIA SANJAYA, JAP LUCY, REINHARD PINONTOAN Department of Biology, Faculty of Science and Technology, Universitas Pelita Harapan. Jl. M.H. Thamrin Boulevard 1100, Lippo Karawaci, Tangerang 15811, Banten, Indonesia. Tel./Fax. +62-21-5460901, email: [email protected] Manuscript received: 11 December 2019. Revision accepted: 20 February 2020. Abstract. Michelle, Siregar RAN, Sanjaya A, Jap L, Pinontoan R. 2020. Methylene blue decolorizing bacteria isolated from water sewage in Yogyakarta, Indonesia. Biodiversitas 21: 1136-1141. The textile industry contributes to water pollution issues all over the world. One of the most commonly applied cationic dye in the textile industry is methylene blue. This study aimed to isolate bacteria with the potential to decolorize methylene blue from dye contaminated sewage water located in Kulon Progo District, Yogyakarta, where several textile industries within the proximity, are located. Characterizations of bacterial candidates were done morphologically and biochemically. Molecular identification was conducted by 16S rRNA sequencing. The ability of isolates to decolorize methylene blue was observed by the reduction of methylene blue’s maximum absorption at the wavelength of 665 nm. The results showed that isolates were identified as Comamonas aquatica and Ralstonia mannitolilytica. C. aquatica PMB-1 and R. mannitolilytica PMB-2 isolates were able to decolorize methylene blue with decolorization percentage of 67.9% and 60.3%, respectively when incubated for 96 hours at 37°C. -

Food Microbiology
Food Microbiology Food Water Dairy Beverage Online Ordering Available Food, Water, Dairy, & Beverage Microbiology Table of Contents 1 Environmental Monitoring Contact Plates 3 Petri Plates 3 Culture Media for Air Sampling 4 Environmental Sampling Boot Swabs 6 Environmental Testing Swabs 8 Surface Sanitizers 8 Hand Sanitation 9 Sample Preparation - Dilution Vials 10 Compact Dry™ 12 HardyCHROM™ Chromogenic Culture Media 15 Prepared Media 24 Agar Plates for Membrane Filtration 26 CRITERION™ Dehydrated Culture Media 28 Pathogen Detection Environmental With Monitoring Contact Plates Baird Parker Agar Friction Lid For the selective isolation and enumeration of coagulase-positive staphylococci (Staphylococcus aureus) on environmental surfaces. HardyCHROM™ ECC 15x60mm contact plate, A chromogenic medium for the detection, 10/pk ................................................................................ 89407-364 differentiation, and enumeration of Escherichia coli and other coliforms from environmental surfaces (E. coli D/E Neutralizing Agar turns blue, coliforms turn red). For the enumeration of environmental organisms. 15x60mm plate contact plate, The media is able to neutralize most antiseptics 10/pk ................................................................................ 89407-354 and disinfectants that may inhibit the growth of environmental organisms. Malt Extract 15x60mm contact plate, Malt Extract is recommended for the cultivation and 10/pk ................................................................................89407-482 -

Prepared Culture Media
PREPARED CULTURE MEDIA 121517SS PREPARED CULTURE MEDIA Made in the USA AnaeroGRO™ DuoPak A 02 Bovine Blood Agar, 5%, with Esculin 13 AnaeroGRO™ DuoPak B 02 Bovine Blood Agar, 5%, with Esculin/ AnaeroGRO™ BBE Agar 03 MacConkey Biplate 13 AnaeroGRO™ BBE/PEA 03 Bovine Selective Strep Agar 13 AnaeroGRO™ Brucella Agar 03 Brucella Agar with 5% Sheep Blood, Hemin, AnaeroGRO™ Campylobacter and Vitamin K 13 Selective Agar 03 Brucella Broth with 15% Glycerol 13 AnaeroGRO™ CCFA 03 Brucella with H and K/LKV Biplate 14 AnaeroGRO™ Egg Yolk Agar, Modified 03 Buffered Peptone Water 14 AnaeroGRO™ LKV Agar 03 Buffered Peptone Water with 1% AnaeroGRO™ PEA 03 Tween® 20 14 AnaeroGRO™ MultiPak A 04 Buffered NaCl Peptone EP, USP 14 AnaeroGRO™ MultiPak B 04 Butterfield’s Phosphate Buffer 14 AnaeroGRO™ Chopped Meat Broth 05 Campy Cefex Agar, Modified 14 AnaeroGRO™ Chopped Meat Campy CVA Agar 14 Carbohydrate Broth 05 Campy FDA Agar 14 AnaeroGRO™ Chopped Meat Campy, Blood Free, Karmali Agar 14 Glucose Broth 05 Cetrimide Select Agar, USP 14 AnaeroGRO™ Thioglycollate with Hemin and CET/MAC/VJ Triplate 14 Vitamin K (H and K), without Indicator 05 CGB Agar for Cryptococcus 14 Anaerobic PEA 08 Chocolate Agar 15 Baird-Parker Agar 08 Chocolate/Martin Lewis with Barney Miller Medium 08 Lincomycin Biplate 15 BBE Agar 08 CompactDry™ SL 16 BBE Agar/PEA Agar 08 CompactDry™ LS 16 BBE/LKV Biplate 09 CompactDry™ TC 17 BCSA 09 CompactDry™ EC 17 BCYE Agar 09 CompactDry™ YMR 17 BCYE Selective Agar with CAV 09 CompactDry™ ETB 17 BCYE Selective Agar with CCVC 09 CompactDry™ YM 17 BCYE -

Microbial Ecology of Denitrification in Biological Wastewater Treatment
water research 64 (2014) 237e254 Available online at www.sciencedirect.com ScienceDirect journal homepage: www.elsevier.com/locate/watres Review Microbial ecology of denitrification in biological wastewater treatment * ** Huijie Lu a, , Kartik Chandran b, , David Stensel c a Department of Civil and Environmental Engineering, University of Illinois at Urbana Champaign, 205 N Mathews, Urbana, IL 61801, USA b Department of Earth and Environmental Engineering, Columbia University, 500 West 120th Street, New York, NY 10027, USA c Department of Civil and Environmental Engineering, University of Washington, Seattle, WA 98195, USA article info abstract Article history: Globally, denitrification is commonly employed in biological nitrogen removal processes to Received 21 December 2013 enhance water quality. However, substantial knowledge gaps remain concerning the overall Received in revised form community structure, population dynamics and metabolism of different organic carbon 26 June 2014 sources. This systematic review provides a summary of current findings pertaining to the Accepted 29 June 2014 microbial ecology of denitrification in biological wastewater treatment processes. DNA Available online 11 July 2014 fingerprinting-based analysis has revealed a high level of microbial diversity in denitrifica- tion reactors and highlighted the impacts of carbon sources in determining overall deni- Keywords: trifying community composition. Stable isotope probing, fluorescence in situ hybridization, Wastewater denitrification microarrays and meta-omics further -

Identification of N2-Fixing Plant-And Fungus-Associated Azoarcus
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Nov. 1997, p. 4331–4339 Vol. 63, No. 11 0099-2240/97/$04.0010 Copyright © 1997, American Society for Microbiology Identification of N2-Fixing Plant- and Fungus-Associated Azoarcus Species by PCR-Based Genomic Fingerprints THOMAS HUREK, BIANCA WAGNER, AND BARBARA REINHOLD-HUREK* Max-Planck-Institut fu¨r Terrestrische Mikrobiologie, Arbeitsgruppe Symbioseforschung, D-35043 Marburg, Germany Received 14 February 1997/Accepted 30 August 1997 Most species of the diazotrophic Proteobacteria Azoarcus spp. occur in association with grass roots, while A. tolulyticus and A. evansii are soil bacteria not associated with a plant host. To facilitate species identification and strain comparison, we developed a protocol for PCR-generated genomic fingerprints, using an automated sequencer for fragment analysis. Commonly used primers targeted to REP (repetitive extragenic palindromic) and ERIC (enterobacterial repetitive intergenic consensus) sequence elements failed to amplify fragments from the two species tested. In contrast, the BOX-PCR assay (targeted to repetitive intergenic sequence elements of Streptococcus) yielded species-specific genomic fingerprints with some strain-specific differences. PCR profiles of an additional PCR assay using primers targeted to tRNA genes (tDNA-PCR, for tRNAIle) were more discriminative, allowing differentiation at species-specific (for two species) or infraspecies-specific level. Our protocol of several consecutive PCR assays consisted of 16S ribosomal DNA (rDNA)-targeted, genus- specific -

22092 Tryptic Soy Broth (TSB, (Tryptone Soya Broth, CASO Broth, Soybean Casein Digest Broth, Casein Soya Broth)
22092 Tryptic Soy Broth (TSB, (Tryptone Soya Broth, CASO Broth, Soybean Casein digest Broth, Casein Soya Broth) The medium will support a luxuriant growth of many fastidious organisms without the addition of serum. Used for confirmation of Campylobacter jejuni by means of the motility test. Composition: Ingredients Grams/Litre Casein peptone (pancreatic) 17.0 Soya peptone (papain digest.) 3.0 Sodium chloride 5.0 Dipotassium hydrogen phosphate 2.5 Glucose 2.5 Final pH 7.3 +/- 0.2 at 25°C Store prepared media below 8°C, protected from direct light. Store dehydrated powder, in a dry place, in tightly-sealed containers at 2-25°C. Directions : Suspend 30 g of dehydrated media in 1 litre of purified filtered water. Sterilize at 121°C for 15 minutes. Cool to 45- 50°C. Mix gently and dispense into sterile Petri dishes or sterile culture tubes. Principle and Interpretation: Casein peptone and Soya peptone provide nitrogen, vitamins and minerals. The natural sugars from Soya peptone and Glucose promote organism growth. Sodium chloride is for the osmotic balance, while Dipotassium hydrogen phosphate is a buffering agent. Tryptone Soya Broth is often for the tube dilution method of antibiotic susceptibility testing. The addition of a small amount of agar ( approx. 0.05-0.2% 05040, add before sterilisation) renders the broth suitable for the cultivation of obligatory anaerobes, such as Clostridium species. The superior growth-promoting properties of Tryptic Soy Broth make it especially useful for the isolation of organisms from blood or other body fluids. Anticoagulants such as sodium polyanetholesulfonate (81305) or sodium citrate (71635) may be added to the broth prior to sterilisation.