Odorant and Taste Receptors Beyond the Nose and Mouth
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Homozygous Deletion of Six Olfactory Receptor Genes in a Subset of Individuals with Beta-Thalassemia
Homozygous Deletion of Six Olfactory Receptor Genes in a Subset of Individuals with Beta-Thalassemia Jessica Van Ziffle, Wendy Yang, Farid F. Chehab* Department of Laboratory Medicine, University of California San Francisco, San Francisco, California, United States of America Abstract Progress in the functional studies of human olfactory receptors has been largely hampered by the lack of a reliable experimental model system. Although transgenic approaches in mice could characterize the function of individual olfactory receptors, the presence of over 300 functional genes in the human genome becomes a daunting task. Thus, the characterization of individuals with a genetic susceptibility to altered olfaction coupled with the absence of particular olfactory receptor genes will allow phenotype/genotype correlations and vindicate the function of specific olfactory receptors with their cognate ligands. We characterized a 118 kb b-globin deletion and found that its 39 end breakpoint extends to the neighboring olfactory receptor region downstream of the b-globin gene cluster. This deletion encompasses six contiguous olfactory receptor genes (OR51V1, OR52Z1, OR51A1P, OR52A1, OR52A5, and OR52A4) all of which are expressed in the brain. Topology analysis of the encoded proteins from these olfactory receptor genes revealed that OR52Z1, OR52A1, OR52A5, and OR52A4 are predicted to be functional receptors as they display integral characteristics of G- proteins coupled receptors. Individuals homozygous for the 118 kb b-globin deletion are afflicted with b-thalassemia due to a homozygous deletion of the b-globin gene and have no alleles for the above mentioned olfactory receptors genes. This is the first example of a homozygous deletion of olfactory receptor genes in human. -

Genetic Variation Across the Human Olfactory Receptor Repertoire Alters Odor Perception
bioRxiv preprint doi: https://doi.org/10.1101/212431; this version posted November 1, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Genetic variation across the human olfactory receptor repertoire alters odor perception Casey Trimmer1,*, Andreas Keller2, Nicolle R. Murphy1, Lindsey L. Snyder1, Jason R. Willer3, Maira Nagai4,5, Nicholas Katsanis3, Leslie B. Vosshall2,6,7, Hiroaki Matsunami4,8, and Joel D. Mainland1,9 1Monell Chemical Senses Center, Philadelphia, Pennsylvania, USA 2Laboratory of Neurogenetics and Behavior, The Rockefeller University, New York, New York, USA 3Center for Human Disease Modeling, Duke University Medical Center, Durham, North Carolina, USA 4Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, North Carolina, USA 5Department of Biochemistry, University of Sao Paulo, Sao Paulo, Brazil 6Howard Hughes Medical Institute, New York, New York, USA 7Kavli Neural Systems Institute, New York, New York, USA 8Department of Neurobiology and Duke Institute for Brain Sciences, Duke University Medical Center, Durham, North Carolina, USA 9Department of Neuroscience, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA *[email protected] ABSTRACT The human olfactory receptor repertoire is characterized by an abundance of genetic variation that affects receptor response, but the perceptual effects of this variation are unclear. To address this issue, we sequenced the OR repertoire in 332 individuals and examined the relationship between genetic variation and 276 olfactory phenotypes, including the perceived intensity and pleasantness of 68 odorants at two concentrations, detection thresholds of three odorants, and general olfactory acuity. -
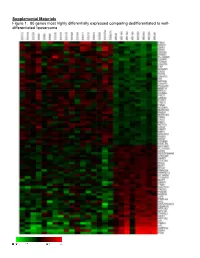
Supplemental Materials Figure 1. 80 Genes Most Highly Differentially Expressed Comparing Dedifferentiated to Well- Differentiated Liposarcoma
Supplemental Materials Figure 1. 80 genes most highly differentially expressed comparing dedifferentiated to well- differentiated liposarcoma Figure 2. Validation of microa CDC2, RACGAP1, FGFR-2, MAD2 and CITED1 200 CDK4 by Liposarcoma Subtype 16 0 12 0 80 Expression40 Level 0 Nl Fat rray data by quantitative RT-P Well Diff 16 0 p16 by Liposarcoma Subtype Dediff 12 0 80 Myxoid Expression40 Level Round Cell 0 12 0 MDM2 by Liposarcoma Subtype 10 0 Pleomorphic Nl Fat 80 60 40 Expression Level 20 RACGAP1 by LiposarcomaWell Diff Subtype 0 70 CR for genes CDK4, MDM2, p16, 60 Dediff 50 Nl Fat 40 30 Myxoid Expression20 Level Well Diff 10 50 0 Round Cell CDC2 by Liposarcoma Subtype Dediff 40 Nl Fat Pleomorphic 30 Myxoid 20 Expression Level Well Diff 10 MAD2L1 by Liposarcoma Subtype Round Cell 60 0 50 Dediff Pleomorphic 40 Nl Fat 30 Myxoid 20 Expression Level Well Diff 10 FGFR-2 by Liposarcoma Subtype 0 Round Cell 200 16 0 Dediff Nl Fat Pleomorphic 12 0 Myxoid 80 Expression Level Well Diff 40 Round Cell 0 Dediff Pleomorphic Nl Fat Myxoid Well Diff CITED1 by Liposarcoma Subtype Round Cell 10 0 80 Dediff Pleomorphic 60 40 Myxoid Expression Level 20 0 Round Cell Nl Fat Pleomorphic Well Diff Dediff Myxoid Round Cell Pleomorphic Table 1. 142 Gene classifier for liposarcoma subtype The genes used for each pair wise subtype comparison are grouped together. The flag column indicates which genes are unique to each subtype comparison. The values show the mean expression levels (actually the mean of the log expression levels was computed and than transformed back to absolute expression level). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Potential Metabolic Effect of Sucralose Following an Oral Glucose Load in Subjects with Obesity and Normal-Weight Subjects
POTENTIAL METABOLIC EFFECT OF SUCRALOSE FOLLOWING AN ORAL GLUCOSE LOAD IN SUBJECTS WITH OBESITY AND NORMAL-WEIGHT SUBJECTS BY ALEXANDER DANIEL NICHOL THESIS Submitted in partial fulfillment of the requirements for the degree of Master of Science in Food Science and Human Nutrition with a concentration in Human Nutrition in the Graduate College of the University of Illinois at Urbana-Champaign, 2018 Urbana, Illinois Master’s Committee: Assistant Professor M. Yanina Pepino de Gruev, Chair Assistant Professor Megan J. Dailey Professor Emeritus John W. Erdman ABSTRACT Objective: Whether sucralose, the most commonly used non-nutritive sweetener (NNS), affects glucose metabolism in people is unclear. It has been reported that, when consumed acutely before an oral glucose tolerance test (OGTT), sucralose enhances insulinemic responses and decreases insulin sensitivity in subjects with obesity who are not regular consumers of NNS. However, studies in normal-weight adults, none of which control for use of NNS, found sucralose does not affect insulin responses to the ingestion of glucose or other carbohydrates. The objectives of the current study are to determine if those effects of sucralose can be replicated in subjects with obesity, are generalizable to normal-weight subjects when controlling for history of NNS use, and are caused merely by the sweet taste of sucralose (i.e., sham-feeding). In addition, with the aim of identifying potential mechanisms by which sucralose may decrease postprandial insulin sensitivity, we here investigated whole-body glucose kinetics by using a dual-tracer approach. Finally, we tested the hypothesis that, due to the compromised intestinal permeability associated with obesity, sucralose consumption is associated with higher plasma sucralose concentrations in people with obesity. -

Sweetness Perception Is Not Involved in the Regulation of Blood Glucose After Oral Application of Sucrose and Glucose Solutions in Healthy Male Subjects
RESEARCH ARTICLE www.mnf-journal.com Sweetness Perception is not Involved in the Regulation of Blood Glucose after Oral Application of Sucrose and Glucose Solutions in Healthy Male Subjects Verena Grüneis, Kerstin Schweiger, Claudia Galassi, Corinna M. Karl, Julia Treml, Jakob P. Ley, Jürgen König, Gerhard E. Krammer, Veronika Somoza, and Barbara Lieder* 1. Introduction Scope: This study investigates the effect of the sweetness of a sucrose versus an isocaloric glucose solution in dietary concentrations on blood glucose Sweet taste is innately highly preferred regulation by adjusting the sweetness level using the sweet taste inhibitor by humans, but the global excessive con- sumption of sweet tasting carbohydrates lactisole. largely contributes to the overall energy Methods and Results: A total of 27 healthy males participated in this intake,[1] leading to an increased risk randomized, crossover study with four treatments: 10% glucose, 10% for obesity and comorbidities like type sucrose, 10% sucrose + 60 ppm lactisole, and 10% glucose + 60 ppm 2 diabetes.[2] One of the main sources lactisole. Plasma glucose, insulin, glucagon-like peptide 1, and glucagon for dietary sugars are sugar-sweetened beverages like fruit drinks, lemonades, levels are measured at baseline and 15, 30, 60, 90, and 120 min after beverage and ice tea.[3] Beside the caloric load, the consumption. Test subjects rated the sucrose solution to be sweeter than the consumption of such sugar-sweetened isocaloric glucose solution, whereas no difference in sweetness is reported beverages is associated with the expo- after addition of lactisole to the sucrose solution. Administration of the less sure to a high level of sweetness. -

The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid
The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome Annette K. Brenner, Elise Aasebø, Maria Hernandez-Valladares, Frode Selheim, Frode Berven, Ida-Sofie Grønningsæter, Sushma Bartaula-Brevik and Øystein Bruserud Supplementary Material S2 of S31 Table S1. Detailed information about the 68 AML patients included in the study. # of blasts Viability Proliferation Cytokine Viable cells Change in ID Gender Age Etiology FAB Cytogenetics Mutations CD34 Colonies (109/L) (%) 48 h (cpm) secretion (106) 5 weeks phenotype 1 M 42 de novo 241 M2 normal Flt3 pos 31.0 3848 low 0.24 7 yes 2 M 82 MF 12.4 M2 t(9;22) wt pos 81.6 74,686 low 1.43 969 yes 3 F 49 CML/relapse 149 M2 complex n.d. pos 26.2 3472 low 0.08 n.d. no 4 M 33 de novo 62.0 M2 normal wt pos 67.5 6206 low 0.08 6.5 no 5 M 71 relapse 91.0 M4 normal NPM1 pos 63.5 21,331 low 0.17 n.d. yes 6 M 83 de novo 109 M1 n.d. wt pos 19.1 8764 low 1.65 693 no 7 F 77 MDS 26.4 M1 normal wt pos 89.4 53,799 high 3.43 2746 no 8 M 46 de novo 26.9 M1 normal NPM1 n.d. n.d. 3472 low 1.56 n.d. no 9 M 68 MF 50.8 M4 normal D835 pos 69.4 1640 low 0.08 n.d. -

A Rose Extract Protects the Skin Against Stress Mediators: a Potential Role of Olfactory Receptors
Supplementary data A Rose extract protects the skin against stress mediators: a potential role of olfactory receptors Romain Duroux 1,*, Anne Mandeau 1, Gaelle Guiraudie-Capraz 2, Yannick Quesnel 3 and Estelle Loing 4 1 IFF-Lucas Meyer Cosmetics, Toulouse, France 2 Aix-Marseille University, CNRS, INP, Marseille, France 3 ChemCom S.A., Brussels, Belgium 4 IFF-Lucas Meyer Cosmetics, Quebec, Canada * Correspondence: [email protected] Supplemental methods Keratinocytes cell culture and skin explants Skin organ cultures were prepared from tissues samples derived from volunteers undergoing routine therapeutic procedures, in collaboration with Alphenyx (Marseille). For this study, skin samples were derived from abdominal or breast tissues of healthy women (30-40 years old). Samples were received 1 day post-surgery and used directly for microdissection or cell extraction. Microdissected human skin was cultured at 37°C under 5% CO2, in DMEM (Dutscher, #L0060-500) plus 10% FBS (Sigma Aldrich, #F7524), 1% Penicillin/Streptomycin (Sigma Aldrich, #P0781-100), and 1% of minimum medium non-essential amino acids 10X (Gibco, #11140-035). For keratinocyte culture, cells were isolated from human skin epidermis. Briefly, skin samples were cut into small pieces and immersed in Thermolysin (Sigma Aldrich, #T7902-100mg) at 4°C overnight. Epidermis was then carefully detached from dermis before being incubated with Trypsin-EDTA for 15 minutes at 37°C. Trypsin inhibitor was next added, the mixture centrifuged and the supernatant discarded before adding KGM2 Medium (Promocell, #C- 20011) with 1% Penicillin/Streptomycin (Sigma Aldrich, #P0781-100). NHEK were then maintained in culture at 37 °C under 5 % CO2 and 95 % relative humidity. -

An Evolutionary Based Strategy for Predicting Rational Mutations in G Protein-Coupled Receptors
Ecology and Evolutionary Biology 2021; 6(3): 53-77 http://www.sciencepublishinggroup.com/j/eeb doi: 10.11648/j.eeb.20210603.11 ISSN: 2575-3789 (Print); ISSN: 2575-3762 (Online) An Evolutionary Based Strategy for Predicting Rational Mutations in G Protein-Coupled Receptors Miguel Angel Fuertes*, Carlos Alonso Department of Microbiology, Centre for Molecular Biology “Severo Ochoa”, Spanish National Research Council and Autonomous University, Madrid, Spain Email address: *Corresponding author To cite this article: Miguel Angel Fuertes, Carlos Alonso. An Evolutionary Based Strategy for Predicting Rational Mutations in G Protein-Coupled Receptors. Ecology and Evolutionary Biology. Vol. 6, No. 3, 2021, pp. 53-77. doi: 10.11648/j.eeb.20210603.11 Received: April 24, 2021; Accepted: May 11, 2021; Published: July 13, 2021 Abstract: Capturing conserved patterns in genes and proteins is important for inferring phenotype prediction and evolutionary analysis. The study is focused on the conserved patterns of the G protein-coupled receptors, an important superfamily of receptors. Olfactory receptors represent more than 2% of our genome and constitute the largest family of G protein-coupled receptors, a key class of drug targets. As no crystallographic structures are available, mechanistic studies rely on the use of molecular dynamic modelling combined with site-directed mutagenesis data. In this paper, we hypothesized that human-mouse orthologs coding for G protein-coupled receptors maintain, at speciation events, shared compositional structures independent, to some extent, of their percent identity as reveals a method based in the categorization of nucleotide triplets by their gross composition. The data support the consistency of the hypothesis, showing in ortholog G protein-coupled receptors the presence of emergent shared compositional structures preserved at speciation events. -

The Odorant Receptor OR2W3 on Airway Smooth Muscle Evokes Bronchodilation Via a Cooperative Chemosensory Tradeoff Between TMEM16A and CFTR
The odorant receptor OR2W3 on airway smooth muscle evokes bronchodilation via a cooperative chemosensory tradeoff between TMEM16A and CFTR Jessie Huanga,1,2, Hong Lama,1, Cynthia Koziol-Whiteb,c, Nathachit Limjunyawongd, Donghwa Kime, Nicholas Kimb, Nikhil Karmacharyac, Premraj Rajkumarf, Danielle Firera, Nicholas M. Dalesiog, Joseph Judec, Richard C. Kurtenh, Jennifer L. Pluznickf, Deepak A. Deshpandei, Raymond B. Penni, Stephen B. Liggette,j, Reynold A. Panettieri Jrc, Xinzhong Dongd,k, and Steven S. Anb,c,2 aDepartment of Environmental Health and Engineering, The Johns Hopkins University Bloomberg School of Public Health, Baltimore, MD 21205; bDepartment of Pharmacology, Rutgers-Robert Wood Johnson Medical School, The State University of New Jersey, Piscataway, NJ 08854; cRutgers Institute for Translational Medicine and Science, New Brunswick, NJ 08901; dSolomon H. Snyder Department of Neuroscience, The Johns Hopkins University School of Medicine, Baltimore, MD 21205; eCenter for Personalized Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL 33612; fDepartment of Physiology, The Johns Hopkins University School of Medicine, Baltimore, MD 21205; gDepartment of Anesthesiology and Critical Care Medicine, The Johns Hopkins University School of Medicine, Baltimore, MD 21205; hDepartment of Physiology and Biophysics, University of Arkansas for Medical Sciences, Little Rock, AR 72205; iDivision of Pulmonary and Critical Care Medicine, Department of Medicine, Center for Translational Medicine, Jane and Leonard Korman -

Misexpression of Cancer/Testis (Ct) Genes in Tumor Cells and the Potential Role of Dream Complex and the Retinoblastoma Protein Rb in Soma-To-Germline Transformation
Michigan Technological University Digital Commons @ Michigan Tech Dissertations, Master's Theses and Master's Reports 2019 MISEXPRESSION OF CANCER/TESTIS (CT) GENES IN TUMOR CELLS AND THE POTENTIAL ROLE OF DREAM COMPLEX AND THE RETINOBLASTOMA PROTEIN RB IN SOMA-TO-GERMLINE TRANSFORMATION SABHA M. ALHEWAT Michigan Technological University, [email protected] Copyright 2019 SABHA M. ALHEWAT Recommended Citation ALHEWAT, SABHA M., "MISEXPRESSION OF CANCER/TESTIS (CT) GENES IN TUMOR CELLS AND THE POTENTIAL ROLE OF DREAM COMPLEX AND THE RETINOBLASTOMA PROTEIN RB IN SOMA-TO- GERMLINE TRANSFORMATION", Open Access Master's Thesis, Michigan Technological University, 2019. https://doi.org/10.37099/mtu.dc.etdr/933 Follow this and additional works at: https://digitalcommons.mtu.edu/etdr Part of the Cancer Biology Commons, and the Cell Biology Commons MISEXPRESSION OF CANCER/TESTIS (CT) GENES IN TUMOR CELLS AND THE POTENTIAL ROLE OF DREAM COMPLEX AND THE RETINOBLASTOMA PROTEIN RB IN SOMA-TO-GERMLINE TRANSFORMATION By Sabha Salem Alhewati A THESIS Submitted in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE In Biological Sciences MICHIGAN TECHNOLOGICAL UNIVERSITY 2019 © 2019 Sabha Alhewati This thesis has been approved in partial fulfillment of the requirements for the Degree of MASTER OF SCIENCE in Biological Sciences. Department of Biological Sciences Thesis Advisor: Paul Goetsch. Committee Member: Ebenezer Tumban. Committee Member: Zhiying Shan. Department Chair: Chandrashekhar Joshi. Table of Contents List of figures .......................................................................................................................v -

Research Article the Differentially Expressed
Ashdin Publishing Journal of Drug and Alcohol Research Vol. 10 (2021), Article ID 236125, 5 pages Research Article The Differentially Expressed Genes and Biomarker Identification for Dengue Disease Using Transcriptome Data Analysis Sunil Krishnan G, Amit Joshi and Vikas Kaushik* Department of Bioinformatics, Lovely Professional University, Punjab, India *Address Correspondence to: Vikas Kaushik, Department of Bioinformatics, Lovely Professional University, Punjab, India, E-mail: [email protected] Received: May 24, 2021; Accepted: June 07, 2021; Published: June 14, 2021 Copyright © 2021 Sunil Krishnan G. This is an open access article distributed under the terms of the Creative Commons Attribution Li- cense, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract [9]. Several microarray studies acknowledged differential- This bioinformatics and biostatistics study was designed to recognize and ly expressed genes (DEGs) from multiple sample profiles examine the differentially expressed genes (DEGs) linked with dengue vi- [10,11]. Consistently DEGs identified from various previ- rus infection in Homo sapiens. Thirty nine transcriptome profile datasets ous studies [12-16] were used to make out a potential bio- were analyzed by linear models for microarray analysis based on the R package of the biostatistics test for the identification of significantly ex- marker for the DENV disease. Meta-analysis approaches pressed genes associated with the disease. The Benjamini and Hochberg are common practice to discover novel DEG signatures for (BH) standard operating procedure assessed DEGs had the least false dis- superior biomarkers and synthetic/biotherapeutics [17,18]. covery rate and chosen for further bioinformatics gene analysis.