Efficient Silencing Gene Construct for Resistance to Multiple Common Bean (Phaseolus Vulgaris L.) Viruses
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Determining the Role of Seed and Soil in the Transmission Of
DETERMINING THE ROLE OF SEED AND SOIL IN THE TRANSMISSION OF VIRUSES CAUSING MAIZE LETHAL NECROSIS DISEASE By GATUNZI KITIRA FELIX (B.Sc. in Rural Development) A thesis submitted in partial fulfillment of requirements for the award of degree of Master of Science in Plant Pathology Department of Plant Science and Crop Protection Faculty of Agriculture University of Nairobi 2018 DECLARATION This thesis is a presentation of my original research work and has not been presented for a degree in any other University. Felix Gatunzi Kitira Date…................................................ This thesis has been submitted with our approval as University supervisors: Dr. Douglas Watuku Miano Department of Plant Science and Crop Protection University of Nairobi Signature ….…………………………. Date……………………………….. Prof. Daniel Mukunya Department of Plant Science and Crop Protection Signature ……………………………. Date……………………………….. Dr. Suresh Lingadahalli Mahabaleswara Global Maize Program (GMP), International Maize and Wheat Improvement Center CIMMYT Signature.…………………………………… Date……………………………… i PLAGIARISM DECLARATION I understand what plagiarism is and I am aware of the University’s policy. In this regard, I declare that this MSc Thesis is my original work. Where other people’s work or my own work has been used, it has properly been acknowledged and referenced in accordance with the University of Nairobi’s requirement. I have not allowed, and shall allow anyone to copy my work with the intention of passing it off as his or her own work. I understand that any false claim in respect of this work shall result in disciplinary action, in accordance with University Plagiarism Policy. Signature.................................................................Date.......................................................... ii DEDICATION To the almighty God for imparting me with his grace along this journey. -

Aphid Transmission of Potyvirus: the Largest Plant-Infecting RNA Virus Genus
Supplementary Aphid Transmission of Potyvirus: The Largest Plant-Infecting RNA Virus Genus Kiran R. Gadhave 1,2,*,†, Saurabh Gautam 3,†, David A. Rasmussen 2 and Rajagopalbabu Srinivasan 3 1 Department of Plant Pathology and Microbiology, University of California, Riverside, CA 92521, USA 2 Department of Entomology and Plant Pathology, North Carolina State University, Raleigh, NC 27606, USA; [email protected] 3 Department of Entomology, University of Georgia, 1109 Experiment Street, Griffin, GA 30223, USA; [email protected] * Correspondence: [email protected]. † Authors contributed equally. Received: 13 May 2020; Accepted: 15 July 2020; Published: date Abstract: Potyviruses are the largest group of plant infecting RNA viruses that cause significant losses in a wide range of crops across the globe. The majority of viruses in the genus Potyvirus are transmitted by aphids in a non-persistent, non-circulative manner and have been extensively studied vis-à-vis their structure, taxonomy, evolution, diagnosis, transmission and molecular interactions with hosts. This comprehensive review exclusively discusses potyviruses and their transmission by aphid vectors, specifically in the light of several virus, aphid and plant factors, and how their interplay influences potyviral binding in aphids, aphid behavior and fitness, host plant biochemistry, virus epidemics, and transmission bottlenecks. We present the heatmap of the global distribution of potyvirus species, variation in the potyviral coat protein gene, and top aphid vectors of potyviruses. Lastly, we examine how the fundamental understanding of these multi-partite interactions through multi-omics approaches is already contributing to, and can have future implications for, devising effective and sustainable management strategies against aphid- transmitted potyviruses to global agriculture. -

Sugarcane Mosaic Virus Infects Stenotaphrum Secundatum in Australia
Australasian Plant Disease Notes (2020) 15:41 https://doi.org/10.1007/s13314-020-00410-y Sugarcane mosaic virus infects Stenotaphrum secundatum in Australia Nga T. Tran1 & Ai Chin Teo1 & John E. Thomas1 & Kathleen S. Crew1,2 & Andrew D. W. Geering1 Received: 28 September 2020 /Accepted: 2 November 2020 # Australasian Plant Pathology Society Inc. 2020 Abstract This study presents the first report of sugarcane mosaic virus (SCMV) infecting Stenotaphrum secundatum (buffalo grass) in Australia, from a turf farm in the Hunter Valley, New South Wales. The plant displayed mosaic symptoms and contained flexuous, filamentous virions of 700–750 × 10–11 nm typical of members of the genus Potyvirus. Infection of the sample by SCMV was confirmed by double antibody sandwich ELISA and RT-PCR amplification of the coat protein coding region of the viral genome. In a phylogenetic analysis, the buffalo grass isolate was sister to a clade of maize-infecting isolates of SCMV from eastern Africa and was 75.8% and 79.4% identical to the exemplar isolate of SCMV at nucleotide and amino acid levels, respectively. Keywords Potyvirus . Buffalo grass . St. Augustinegrass . Nucleotide sequence Stenotaphrum secundatum (buffalo grass in Australia or St. SCMV has a worldwide distribution and infects several Augustinegrass in the USA) is a hardwearing and vigorous economically important monocotyledonous crops including warm season turfgrass species, thought to be endemic to the maize (Zea mays), sorghum (Sorghum bicolor) and sugarcane Atlantic coasts of the Americas and Africa (Sauer 1972; (Saccharum officinarum) (Yang and Mirkov 1997;Wuetal. Busey 2003). It is the most important turf species in 2012). -

Effects of Plant Viruses on Vectors and Non-Vector Herbivores in Three
Louisiana State University LSU Digital Commons LSU Doctoral Dissertations Graduate School March 2019 Effects of Plant Viruses on Vectors and Non-vector Herbivores in Three Different Pathosystems Sunil Paudel Louisiana State University and Agricultural and Mechanical College, [email protected] Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_dissertations Part of the Entomology Commons Recommended Citation Paudel, Sunil, "Effects of Plant Viruses on Vectors and Non-vector Herbivores in Three Different Pathosystems" (2019). LSU Doctoral Dissertations. 4870. https://digitalcommons.lsu.edu/gradschool_dissertations/4870 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Doctoral Dissertations by an authorized graduate school editor of LSU Digital Commons. For more information, please [email protected]. EFFECTS OF PLANT VIRUSES ON VECTORS AND NON-VECTOR HERBIVORES IN THREE DIFFERENT PATHOSYSTEMS A Dissertation Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The Department of Entomology by Sunil Paudel B. S., Tribhuvan University, 2008 M.S., University of Idaho, 2013 May 2019 ACKNOWLEDGEMENTS I would like to express my sincere gratitude to my major advisor, Dr. Jeffrey A. Davis for his continuous support throughout my graduate studies, for his patience, encouragement, and guidance that helped me to grow as a researcher and a critical thinker. I am also thankful to my dissertation committee members, Drs. Michael J. Stout, Fangneng Huang, Jeffrey W. Hoy, and Dean’s representative Dr. -
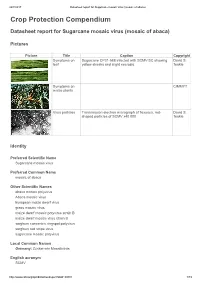
Datasheet Report for Sugarcane Mosaic Virus (Mosaic of Abaca)
24/7/2017 Datasheet report for Sugarcane mosaic virus (mosaic of abaca) Crop Protection Compendium Datasheet report for Sugarcane mosaic virus (mosaic of abaca) Pictures Picture Title Caption Copyright Symptoms on Sugarcane CP31-588 infected with SCMV-SC showing David S. leaf yellow streaks and slight necrosis. Teakle Symptoms on CIMMYT maize plants Virus particles Transmission electron micrograph of flexuous, rod- David S. shaped particles of SCMV x40 000. Teakle Identity Preferred Scientific Name Sugarcane mosaic virus Preferred Common Name mosaic of abaca Other Scientific Names abaca mosaic potyvirus Abaca mosaic virus European maize dwarf virus grass mosaic virus maize dwarf mosaic potyvirus strain B maize dwarf mosaic virus strain B sorghum concentric ringspot potyvirus sorghum red stripe virus sugarcane mosaic potyvirus Local Common Names Germany: Zuckerrohr Mosaikvirus English acronym SCMV http://www.cabi.org/cpc/datasheetreport?dsid=49801 1/19 24/7/2017 Datasheet report for Sugarcane mosaic virus (mosaic of abaca) EPPO code SCMV00 (Sugarcane mosaic potyvirus) Taxonomic Tree Domain: Virus Group: "Positive sense ssRNA viruses" Group: "RNA viruses" Family: Potyviridae Genus: Potyvirus Species: Sugarcane mosaic virus Notes on Taxonomy and Nomenclature This virus was described from sugarcane by Brandes (1919). Strains naturally infecting certain other hosts are also known as abaca mosaic potyvirus (Eloja and Tinsley, 1963) and as maize dwarf mosaic potyvirus strain B (Mackenzie et al., 1966; Louie and Knoke, 1975). Until recently, most aphid-borne potyviruses infecting sugarcane and other members of the Poaceae were assumed to be either sugarcane mosaic virus or maize dwarf mosaic virus. However, new criteria for speciation of these members of the Potyviridae have been proposed (Shukla et al., 1989; Shukla et al., 1992; Shukla and Ward, 1994): the amino-acid sequence homology of their coat proteins, and serological relationships with antisera derived from epitopes in the amino terminus rather than the core region of the coat protein. -

Complete Nucleotide Sequence and Genetic Organization of Sunflower Mild Mosaic Virus (Summov)
Complete nucleotide sequence and genetic organization of Sunflower mild mosaic virus (SuMMoV) Fabián Giolitti1, Soledad de Breuil1, Nicolas Bejerman1 and Sergio Lenardon1,2 [email protected]; [email protected] 1- Instituto de Patología Vegetal (IPAVE), Centro de Investigaciones Agropecuarias (CIAP), Instituto Nacional de Tecnología Agropecuaria (INTA), Camino 60 cuadras Km. 5,5, X5020ICA, Córdoba, Argentina. 2- Facultad de Agronomía y Veterinaria Universidad, Nacional de Río Cuarto. ABSTRACT Two potyviruses that infect sunflower (Helianthus annuus L.) crops have been reported in Argentina. Sunflower chlorotic mottle virus (SuCMoV) is the most widely distributed one, with two strains: common and ring spot, which have been completely characterized. Sunflower mild mosaic virus (SuMMoV) that produces a chlorotic mild mosaic on sunflower commercial crops was detected in the rural area of Paraná (Entre Ríos Province). This virus has been biologically and serologically characterized but not fully characterized at the molecular level. The aim of this work was to determine the complete sequence of SuMMoV and study its genomic organization. A total RNA extraction was made from a preparation enriched with this virus. The quality of preparation was confirmed through electron microscopy and RT-PCR with specific primers for capsid protein gene. Total RNA obtained was pyrosequenced and the sequences obtained were analyzed. Electron microscopy from rapid preparations of samples enriched with the virus showed high concentration of potyvirus-like particles. RT-PCR showed an aplicon of expected size. The pyrosequencing resulted in a large contig of 9686 nucleotides (nt) with a single open reading frame (ORF) of 3077 amino acids (aa). -

Viruses in Maize and Johnsongrass in Southern Ohio
Virology e-Xtra* Viruses in Maize and Johnsongrass in Southern Ohio L. R. Stewart, R. Teplier, J. C. Todd, M. W. Jones, B. J. Cassone, S. Wijeratne, A. Wijeratne, and M. G. Redinbaugh First, third, fourth, fifth, and eighth authors: U.S. Department of Agriculture, Agricultural Research Services, Corn, Soybean and Wheat Quality Research Unit, Wooster, OH; first and eighth authors: The Ohio State University, Department of Plant Pathology, Wooster; second author: University of Avignon, Avignon, France; and sixth and seventh authors: The Ohio State University, Molecular and Cellular Imaging Center, Wooster. Accepted for publication 2 June 2014. ABSTRACT Stewart, L. R., Teplier, R., Todd, J. C., Jones, M. W., Cassone, B. J., viruses present at the sites of past disease emergence, we used a combina- Wijeratne, S., Wijeratne, A., and Redinbaugh, M. G. 2014. Viruses in tion of serological testing and next-generation RNA sequencing approaches. maize and Johnsongrass in southern Ohio. Phytopathology 104:1360- Here, we report enzyme-linked immunosorbent assay and RNA-Seq data 1369. from samples collected over 2 years to assess the presence of viruses in cultivated maize and an important weedy reservoir, Johnsongrass (Sorghum The two major U.S. maize viruses, Maize dwarf mosaic virus halepense). Results revealed a persistent reservoir of MDMV and two (MDMV) and Maize chlorotic dwarf virus (MCDV), emerged in southern strains of MCDV in Ohio Johnsongrass. We identified sequences of Ohio and surrounding regions in the 1960s and caused significant losses. several other grass-infecting viruses and confirmed the presence of Wheat Planting resistant varieties and changing cultural practices has dramati- mosaic virus in Ohio maize. -
Sugarcane Germplasm Conservation and Exchange
Sugarcane Germplasm Conservation and Exchange Report of an international workshop held in Brisbane, Queensland, Australia 28-30 June 1995 Editors B.J. Croft, C.M. Piggin, E.S. Wallis and D.M. Hogarth The Australian Centre for International Agricultural Research (ACIAR) was established in June 1982 by an Act of the Australian Parliament. [ts mandate is to help identify agricultural problems in developing countries and to commission collaborative research between Australian and developing country resean;hers in field, where Australia has special research competence. Where trade names are used thi, does not constitute endorsement of nor discrimination against any product by the Centre. This series of publications includes the full proceedings of research workshops or symposia or supported by ACIAR. Numbers in this ,,,ries are distributed internationally to individuals and scientific institutions. © Australian Centre for International Agricultural Research GPO Box 1571. Canberra, ACT 260 J Australia Croft, B.1., Piggin, CM., Wallis E.S. and Hogarth, D.M. 1996. Sugarcane germplasm conservation and exchange. ACIAR Proceedings No. 67, 134p. ISBN 1 86320 177 7 Editorial management: P.W. Lynch Typesetting and page layout: Judy Fenelon, Bawley Point. New Soulh Wales Printed by: Paragon Printers. Fyshwick, ACT Foreword ANALYSIS by the Technical Advisory Committee of the Consultative Group of Internation al Agricultural Research Centres (estimated that in 1987-88 sugar was the fourteenth most important crop in developing countries, with a gross value of production of over US$7.3 billion. In Australia it is the third most important crop, with a value in 1994-95 of around A$1.7 billion. -

Characterization of a Sorghum Mosaic Virus (Srmv) Isolate in China
Saudi Journal of Biological Sciences (2015) xxx, xxx–xxx King Saud University Saudi Journal of Biological Sciences www.ksu.edu.sa www.sciencedirect.com ORIGINAL ARTICLE Characterization of a Sorghum mosaic virus (SrMV) isolate in China Yu Liang Zhang a,b, Kayla K. Pennerman c, Hongxing Wang a, Guohua Yin c,* a Key Laboratory of Biology and Genetic Resources of Tropical Crops, Ministry of Agriculture, Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, Haikou, Hainan 5711001, China b Sugarcane Research Center, Chinese Academy of Tropical Agricultural Sciences, Haikou, Hainan 5711001, China c Department of Plant Biology and Pathology, Rutgers, The State University of New Jersey, New Brunswick, NJ 08901, United States Received 3 December 2014; revised 5 February 2015; accepted 7 February 2015 KEYWORDS Abstract Sorghum mosaic virus (SrMV), a causal agent of the destructive sugarcane mosaic dis- SrMV; ease, has a global presence. An isolate of SrMV infecting a commercially-grown sugarcane plant Sugarcane mosaic disease; was recovered from the Hainan province of China. The virions were visualized by an electron Coat protein microscope, and the coat proteins (CPs) were sequenced by matrix-assisted laser desorption/ioniza- tion time-of-flight mass spectrometry and tandem mass spectrometry. Discrepancies between the CP predicted and actual amino acid sequences were noted, which confounded the phylogenetic assign- ment of the isolate. The apparent variations may have physiological effects on the pathogenicity and virulence of SrMV. ª 2015 The Authors. Production and hosting by Elsevier B.V. on behalf of King Saud University. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/). -

Abacá Mosaic Virus: a Distinct Strain of Sugarcane Mosaic Virus
CSIRO PUBLISHING www.publish.csiro.au/journals/app Australasian Plant Pathology, 2004, 33, 475–484 Abaca´ mosaic virus: a distinct strain of Sugarcane mosaic virus C. F.GambleyA, J. E. ThomasA,C, L. V.MagnayeB and L. HerraduraB ADepartment of Primary Industries and Fisheries, Horticulture and Forest Sciences, 80 Meiers Road, Indooroopilly, Qld 4068, Australia. BDepartment of Agriculture, Bureau of Plant Industry, Bago-Oshiro 8000, Davao City, Philippines. CCorresponding author. Email: [email protected] Abstract. Abaca´ mosaic virus (AbaMV) is related to members of the sugarcane mosaic virus subgroup of the genus Potyvirus. The ∼2kb3 terminal region of the viral genome was sequenced and, in all areas analysed, found to be most similar to Sugarcane mosaic virus (SCMV) and distinct from Johnsongrass mosaic virus (JGMV), Maize dwarf mosaic virus (MDMV) and Sorghum mosaic virus (SrMV). Cladograms of the 3 terminal region of the NIb protein, the coat protein core and the 3 untranslated region showed that AbaMV clustered with SCMV, which was a distinct clade and separate from JGMV,MDMV and SrMV.The N-terminal region of the AbaMV coat protein had a unique amino acid repeat motif different from those previously published for other strains of SCMV. The first experimental transmission of AbaMV from abaca(´ Musa textilis) to banana (Musa sp.), using the aphid vectors Rhopalosiphum maidis and Aphis gossypii, is reported. Polyclonal antisera for the detection of AbaMV in western blot assays and ELISA were prepared from recombinant coat protein expressed in E. coli. A reverse transcriptase PCR diagnostic assay, with microtitre plate colourimetric detection, was developed to discriminate between AbaMV and Banana bract mosaic virus, another Musa-infecting potyvirus. -

Maize Dwarf Mosaic Virus CP
INDUSTRY BIOSECURITY PLAN FOR THE GRAINS INDUSTRY Threat Specific Contingency Plan Maize dwarf mosaic virus Prepared by Brendan Rodoni and Plant Health Australia January 2011 PLANTH HEALTH AUSTRALIA | Contingency Plan – Maize dwarf mosaic virus Disclaimer The scientific and technical content of this document is current to the date published and all efforts were made to obtain relevant and published information on the pest. New information will be included as it becomes available, or when the document is reviewed. The material contained in this publication is produced for general information only. It is not intended as professional advice on any particular matter. No person should act or fail to act on the basis of any material contained in this publication without first obtaining specific, independent professional advice. Plant Health Australia and all persons acting for Plant Health Australia in preparing this publication, expressly disclaim all and any liability to any persons in respect of anything done by any such person in reliance, whether in whole or in part, on this publication. The views expressed in this publication are not necessarily those of Plant Health Australia. Further information For further information regarding this contingency plan, contact Plant Health Australia through the details below. Address: Suite 5, FECCA House 4 Phipps Close DEAKIN ACT 2600 Phone: +61 2 6215 7700 Fax: +61 2 6260 4321 Email: [email protected] Website: www.planthealthaustralia.com.au PAGE 2 PLANTH HEALTH AUSTRALIA | Contingency Plan – -
Complete Sections As Applicable
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the separate document “Help with completing a taxonomic proposal” Please try to keep related proposals within a single document; you can copy the modules to create more than one genus within a new family, for example. MODULE 1: TITLE, AUTHORS, etc (to be completed by ICTV Code assigned: 2016.008a,bP officers) Short title: Create three species in genus Potyvirus and abolish five species in genus Potyvirus (e.g. 6 new species in the genus Zetavirus) Modules attached 2 3 4 5 (modules 1 and 11 are required) 6 7 8 9 10 Author(s): Wylie, Stephen (Chair) [email protected] Adams, Michael J. [email protected] Chalam, Celia [email protected] Kreuze, Jan F. [email protected] Lopez-Moya, Juan Jose [email protected] Ohshima, Kazusato [email protected] Praveen, Shelly [email protected] Rabenstein, Frank [email protected] Stenger, Drake C. [email protected] Wang, Aiming [email protected] Zerbini, F. Murilo [email protected] Corresponding author with e-mail address: Stephen Wylie [email protected] List the ICTV study group(s) that have seen this proposal: A list of study groups and contacts is provided at http://www.ictvonline.org/subcommittees.asp Potyviridae . If in doubt, contact the appropriate subcommittee chair (fungal, invertebrate, plant, prokaryote or vertebrate viruses) ICTV Study Group comments (if any) and response of the proposer: Date first submitted to ICTV: 2016 Date of this revision (if different to above): Page 1 of 10 ICTV-EC comments and response of the proposer: EC comment: SG response: Page 2 of 10 MODULE 2: NEW SPECIES creating and naming one or more new species.