Cryptic Organelles in Parasitic Protists and Fungi
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Download the Abstract Book
1 Exploring the male-induced female reproduction of Schistosoma mansoni in a novel medium Jipeng Wang1, Rui Chen1, James Collins1 1) UT Southwestern Medical Center. Schistosomiasis is a neglected tropical disease caused by schistosome parasites that infect over 200 million people. The prodigious egg output of these parasites is the sole driver of pathology due to infection. Female schistosomes rely on continuous pairing with male worms to fuel the maturation of their reproductive organs, yet our understanding of their sexual reproduction is limited because egg production is not sustained for more than a few days in vitro. Here, we explore the process of male-stimulated female maturation in our newly developed ABC169 medium and demonstrate that physical contact with a male worm, and not insemination, is sufficient to induce female development and the production of viable parthenogenetic haploid embryos. By performing an RNAi screen for genes whose expression was enriched in the female reproductive organs, we identify a single nuclear hormone receptor that is required for differentiation and maturation of germ line stem cells in female gonad. Furthermore, we screen genes in non-reproductive tissues that maybe involved in mediating cell signaling during the male-female interplay and identify a transcription factor gli1 whose knockdown prevents male worms from inducing the female sexual maturation while having no effect on male:female pairing. Using RNA-seq, we characterize the gene expression changes of male worms after gli1 knockdown as well as the female transcriptomic changes after pairing with gli1-knockdown males. We are currently exploring the downstream genes of this transcription factor that may mediate the male stimulus associated with pairing. -

Autophagy in Trypanosomatids
Cells 2012, 1, 346-371; doi:10.3390/cells1030346 OPEN ACCESS cells ISSN 2073-4409 www.mdpi.com/journal/cells Review Autophagy in Trypanosomatids Ana Brennand 1,†, Eva Rico 2,†,‡ and Paul A. M. Michels 1,* 1 Research Unit for Tropical Diseases, de Duve Institute, Université catholique de Louvain, Avenue Hippocrate 74, postal box B1.74.01, B-1200 Brussels, Belgium; E-Mail: [email protected] 2 Department of Biochemistry and Molecular Biology, University Campus, University of Alcalá, Alcalá de Henares, Madrid, 28871, Spain; E-Mail: [email protected] † These authors contributed equally to this work. ‡ Present Address: Centre for Immunity, Infection and Evolution, Institute of Immunology and Infection Research, School of Biological Sciences, King’s Buildings, University of Edinburgh, West Mains Road, Edinburgh EH9 3JT, UK. * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +32-2-7647473; Fax: +32-2-7626853. Received: 28 June 2012; in revised form: 14 July 2012 / Accepted: 16 July 2012 / Published: 27 July 2012 Abstract: Autophagy is a ubiquitous eukaryotic process that also occurs in trypanosomatid parasites, protist organisms belonging to the supergroup Excavata, distinct from the supergroup Opistokontha that includes mammals and fungi. Half of the known yeast and mammalian AuTophaGy (ATG) proteins were detected in trypanosomatids, although with low sequence conservation. Trypanosomatids such as Trypanosoma brucei, Trypanosoma cruzi and Leishmania spp. are responsible for serious tropical diseases in humans. The parasites are transmitted by insects and, consequently, have a complicated life cycle during which they undergo dramatic morphological and metabolic transformations to adapt to the different environments. -

A Wide Diversity of Previously Undetected Freeliving
Environmental Microbiology (2010) 12(10), 2700–2710 doi:10.1111/j.1462-2920.2010.02239.x A wide diversity of previously undetected free-living relatives of diplomonads isolated from marine/saline habitatsemi_2239 2700..2710 Martin Kolisko,1 Jeffrey D. Silberman,2 Kipferlia n. gen. The remaining isolates include rep- Ivan Cepicka,3 Naoji Yubuki,4† Kiyotaka Takishita,5 resentatives of three other lineages that likely repre- Akinori Yabuki,4 Brian S. Leander,6 Isao Inouye,4 sent additional undescribed genera (at least). Small- Yuji Inagaki,7 Andrew J. Roger8 and subunit ribosomal RNA gene phylogenies show that Alastair G. B. Simpson1* CLOs form a cloud of six major clades basal to the Departments of 1Biology and 8Biochemistry and diplomonad-retortamonad grouping (i.e. each of the Molecular Biology, Dalhousie University, Halifax, Nova six CLO clades is potentially as phylogenetically Scotia, Canada. distinct as diplomonads and retortamonads). CLOs 2Department of Biological Sciences, University of will be valuable for tracing the evolution of Arkansas, Fayetteville, AR, USA. diplomonad cellular features, for example, their 3Department of Zoology, Faculty of Science, Charles extremely reduced mitochondrial organelles. It is University in Prague, Prague, Czech Republic. striking that the majority of CLO diversity was unde- 4Institute of Biological Sciences, Graduate School of Life tected by previous light microscopy surveys and and Environmental Sciences and 7Center for environmental PCR studies, even though they inhabit Computational Sciences and Institute of Biological a commonly sampled environment. There is no Sciences, University of Tsukuba, Tsukuba, Ibaraki, reason to assume this is a unique situation – it is Japan. likely that undersampling at the level of major lin- 5Japan Agency for Marine-Earth Science and eages is still widespread for protists. -
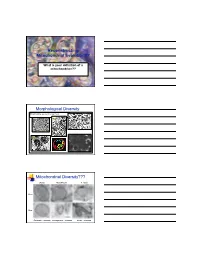
Reconstructing Mitochondrial Evolution?? Morphological Diversity Mitochondrial Diversity???
Reconstructing Mitochondrial Evolution?? What is your definition of a mitochondrion?? Morphological Diversity Mitochondria as we all know them: Suprarenal gland Liver cell Plasma cell Adrenal cortex Mitochondrial Diversity??? Chicken Neocallimastix T. foetus 100 nm 50 nm Entamoeba - mitosome microsporidian - mitosome Giardia - mitosome Reconstructing Evolution Mitochondrial evolution well established endosymbiotic theory α-proteobacterium - Rickettsia prowazekii Hydrogenosomal evolution No DNA NOW 2 examples Nyctotherus and Blastocystis (MLO) Several proteins similar to mitochondria Mitosome evolution No DNA Few proteins identified similar to mitochondria Origins via Endosymbiosis Current dogma - mitochondria and related Aerobic α-proteobacterium prokaryote gave rise to present day mitochondria. organelles arose just once in evolution Are hydrogenosomes and mitosomes of anaerobic protists derived from the same proto-mitochondrion? Evidence for: accumulating evidence for several proteins that are currently found in mitochondria - Proteins of Fe-S cluster formation. Scenario A Common ancestor facultative Scenario B organism? degenerate mitochondrion invoke lateral gene transfer from anaerobic prokaryotes Hypotheses for Mito Acquisition Mitochondrion-related Organelles 2 way to interpret this summary - can you think of both? Groups with mitochondrial homologues Hjort et al Phil. Trans. R. Soc. B 2010 365, 713-727 Origin of Hydrogenosomes (Mitosomes?) 1. a)Conversion of Mitochondria HYD b)Common ancestor with mitochondria ? HYD 2. Independent Origin α-proteobacterium ? - anaerobic bacterium HYD Organelles - origins and biogenesis Approaches: (1) Conduct phylogenetic analyses of similar proteins Hsp70 Fd Hsp60 Isc subunits (2) Examine protein targeting to the organelle matrix protein targeting membrane protein targeting (3) Characterize membrane/translocation components These components could have evolved as the endosymbiont was converted to organelle. Reveals evolutionary history. -

Protist Phylogeny and the High-Level Classification of Protozoa
Europ. J. Protistol. 39, 338–348 (2003) © Urban & Fischer Verlag http://www.urbanfischer.de/journals/ejp Protist phylogeny and the high-level classification of Protozoa Thomas Cavalier-Smith Department of Zoology, University of Oxford, South Parks Road, Oxford, OX1 3PS, UK; E-mail: [email protected] Received 1 September 2003; 29 September 2003. Accepted: 29 September 2003 Protist large-scale phylogeny is briefly reviewed and a revised higher classification of the kingdom Pro- tozoa into 11 phyla presented. Complementary gene fusions reveal a fundamental bifurcation among eu- karyotes between two major clades: the ancestrally uniciliate (often unicentriolar) unikonts and the an- cestrally biciliate bikonts, which undergo ciliary transformation by converting a younger anterior cilium into a dissimilar older posterior cilium. Unikonts comprise the ancestrally unikont protozoan phylum Amoebozoa and the opisthokonts (kingdom Animalia, phylum Choanozoa, their sisters or ancestors; and kingdom Fungi). They share a derived triple-gene fusion, absent from bikonts. Bikonts contrastingly share a derived gene fusion between dihydrofolate reductase and thymidylate synthase and include plants and all other protists, comprising the protozoan infrakingdoms Rhizaria [phyla Cercozoa and Re- taria (Radiozoa, Foraminifera)] and Excavata (phyla Loukozoa, Metamonada, Euglenozoa, Percolozoa), plus the kingdom Plantae [Viridaeplantae, Rhodophyta (sisters); Glaucophyta], the chromalveolate clade, and the protozoan phylum Apusozoa (Thecomonadea, Diphylleida). Chromalveolates comprise kingdom Chromista (Cryptista, Heterokonta, Haptophyta) and the protozoan infrakingdom Alveolata [phyla Cilio- phora and Miozoa (= Protalveolata, Dinozoa, Apicomplexa)], which diverged from a common ancestor that enslaved a red alga and evolved novel plastid protein-targeting machinery via the host rough ER and the enslaved algal plasma membrane (periplastid membrane). -

The Apicoplast: a Review of the Derived Plastid of Apicomplexan Parasites
Curr. Issues Mol. Biol. 7: 57-80. Online journalThe Apicoplastat www.cimb.org 57 The Apicoplast: A Review of the Derived Plastid of Apicomplexan Parasites Ross F. Waller1 and Geoffrey I. McFadden2,* way to apicoplast discovery with studies of extra- chromosomal DNAs recovered from isopycnic density 1Botany, University of British Columbia, 3529-6270 gradient fractionation of total Plasmodium DNA. This University Boulevard, Vancouver, BC, V6T 1Z4, Canada group recovered two DNA forms; one a 6kb tandemly 2Plant Cell Biology Research Centre, Botany, University repeated element that was later identifed as the of Melbourne, 3010, Australia mitochondrial genome, and a second, 35kb circle that was supposed to represent the DNA circles previously observed by microscopists (Wilson et al., 1996b; Wilson Abstract and Williamson, 1997). This molecule was also thought The apicoplast is a plastid organelle, homologous to to be mitochondrial DNA, and early sequence data of chloroplasts of plants, that is found in apicomplexan eubacterial-like rRNA genes supported this organellar parasites such as the causative agents of Malaria conclusion. However, as the sequencing effort continued Plasmodium spp. It occurs throughout the Apicomplexa a new conclusion, that was originally embraced with and is an ancient feature of this group acquired by the some awkwardness (“Have malaria parasites three process of endosymbiosis. Like plant chloroplasts, genomes?”, Wilson et al., 1991), began to emerge. apicoplasts are semi-autonomous with their own genome Gradually, evermore convincing character traits of a and expression machinery. In addition, apicoplasts import plastid genome were uncovered, and strong parallels numerous proteins encoded by nuclear genes. These with plastid genomes from non-photosynthetic plants nuclear genes largely derive from the endosymbiont (Epifagus virginiana) and algae (Astasia longa) became through a process of intracellular gene relocation. -

Molecular Identification and Evolution of Protozoa Belonging to the Parabasalia Group and the Genus Blastocystis
UNIVERSITAR DEGLI STUDI DI SASSARI SCUOLA DI DOTTORATO IN SCIENZE BIOMOLECOLARI E BIOTECNOLOGICHE (Intenational PhD School in Biomolecular and Biotechnological Sciences) Indirizzo: Microbiologia molecolare e clinica Molecular identification and evolution of protozoa belonging to the Parabasalia group and the genus Blastocystis Direttore della scuola: Prof. Masala Bruno Relatore: Prof. Pier Luigi Fiori Correlatore: Dott. Eric Viscogliosi Tesi di Dottorato : Dionigia Meloni XXIV CICLO Nome e cognome: Dionigia Meloni Titolo della tesi : Molecular identification and evolution of protozoa belonging to the Parabasalia group and the genus Blastocystis Tesi di dottorato in scienze Biomolecolari e biotecnologiche. Indirizzo: Microbiologia molecolare e clinica Universit degli studi di Sassari UNIVERSITAR DEGLI STUDI DI SASSARI SCUOLA DI DOTTORATO IN SCIENZE BIOMOLECOLARI E BIOTECNOLOGICHE (Intenational PhD School in Biomolecular and Biotechnological Sciences) Indirizzo: Microbiologia molecolare e clinica Molecular identification and evolution of protozoa belonging to the Parabasalia group and the genus Blastocystis Direttore della scuola: Prof. Masala Bruno Relatore: Prof. Pier Luigi Fiori Correlatore: Dott. Eric Viscogliosi Tesi di Dottorato : Dionigia Meloni XXIV CICLO Nome e cognome: Dionigia Meloni Titolo della tesi : Molecular identification and evolution of protozoa belonging to the Parabasalia group and the genus Blastocystis Tesi di dottorato in scienze Biomolecolari e biotecnologiche. Indirizzo: Microbiologia molecolare e clinica Universit degli studi di Sassari Abstract My thesis was conducted on the study of two groups of protozoa: the Parabasalia and Blastocystis . The first part of my work was focused on the identification, pathogenicity, and phylogeny of parabasalids. We showed that Pentatrichomonas hominis is a possible zoonotic species with a significant potential of transmission by the waterborne route and could be the aetiological agent of gastrointestinal troubles in children. -

The Revised Classification of Eukaryotes
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/231610049 The Revised Classification of Eukaryotes Article in Journal of Eukaryotic Microbiology · September 2012 DOI: 10.1111/j.1550-7408.2012.00644.x · Source: PubMed CITATIONS READS 961 2,825 25 authors, including: Sina M Adl Alastair Simpson University of Saskatchewan Dalhousie University 118 PUBLICATIONS 8,522 CITATIONS 264 PUBLICATIONS 10,739 CITATIONS SEE PROFILE SEE PROFILE Christopher E Lane David Bass University of Rhode Island Natural History Museum, London 82 PUBLICATIONS 6,233 CITATIONS 464 PUBLICATIONS 7,765 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: Biodiversity and ecology of soil taste amoeba View project Predator control of diversity View project All content following this page was uploaded by Smirnov Alexey on 25 October 2017. The user has requested enhancement of the downloaded file. The Journal of Published by the International Society of Eukaryotic Microbiology Protistologists J. Eukaryot. Microbiol., 59(5), 2012 pp. 429–493 © 2012 The Author(s) Journal of Eukaryotic Microbiology © 2012 International Society of Protistologists DOI: 10.1111/j.1550-7408.2012.00644.x The Revised Classification of Eukaryotes SINA M. ADL,a,b ALASTAIR G. B. SIMPSON,b CHRISTOPHER E. LANE,c JULIUS LUKESˇ,d DAVID BASS,e SAMUEL S. BOWSER,f MATTHEW W. BROWN,g FABIEN BURKI,h MICAH DUNTHORN,i VLADIMIR HAMPL,j AARON HEISS,b MONA HOPPENRATH,k ENRIQUE LARA,l LINE LE GALL,m DENIS H. LYNN,n,1 HILARY MCMANUS,o EDWARD A. D. -

Reevaluation of the Toxoplasma Gondii and Neospora Caninum Genomes Reveals Misassembly, Karyotype Differences and Chromosomal Rearrangements
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.22.111195; this version posted May 25, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Reevaluation of the Toxoplasma gondii and Neospora caninum genomes reveals misassembly, karyotype differences and chromosomal rearrangements Luisa Berna1, Pablo Marquez1, Andrés Cabrera1, Gonzalo Greif1, María E. Francia2,3,*, Carlos Robello1,4,* AUTHORS AFFILIATIONS 1 Laboratory of Host Pathogen Interactions-Molecular Biology Unit. Institut Pasteur de Montevideo. Montevideo, Uruguay 2 Laboratory of Apicomplexan Biology. Institut Pasteur de Montevideo. Montevideo, Uruguay. 3 Departamento de Parasitología y Micología. Facultad de Medicina-Universidad de la República. Montevideo, Uruguay. 4 Departamento de Bioquímica. Facultad de Medicina-Universidad de la República. Montevideo, Uruguay. *Corresponding authors ABSTRACT Neospora caninum primarily infects cattle causing abortions with an estimated impact of a billion dollars on worldwide economy, annually. However, the study of its biology has been unheeded by the established paradigm that it is virtually identical to its close relative, the widely studied human pathogen, Toxoplasma gondii. By revisiting the genome sequence, assembly and annotation using third generation sequencing technologies, here we show that the N. caninum genome was originally incorrectly assembled under the presumption of synteny with T. gondii. We show that major chromosomal rearrangements have occurred between these species. Importantly, we show that chromosomes originally annotated as ChrVIIb and VIII are indeed fused, reducing the karyotype of both N. -

The NTP Generating Activity of Pyruvate Kinase II Is Critical
RESEARCH ARTICLE The NTP generating activity of pyruvate kinase II is critical for apicoplast maintenance in Plasmodium falciparum Russell P Swift, Krithika Rajaram, Cyrianne Keutcha, Hans B Liu, Bobby Kwan, Amanda Dziedzic, Anne E Jedlicka, Sean T Prigge* Department of Molecular Microbiology and Immunology, Johns Hopkins Bloomberg School of Public Health, Baltimore, United States Abstract The apicoplast of Plasmodium falciparum parasites is believed to rely on the import of three-carbon phosphate compounds for use in organelle anabolic pathways, in addition to the generation of energy and reducing power within the organelle. We generated a series of genetic deletions in an apicoplast metabolic bypass line to determine which genes involved in apicoplast carbon metabolism are required for blood-stage parasite survival and organelle maintenance. We found that pyruvate kinase II (PyrKII) is essential for organelle maintenance, but that production of pyruvate by PyrKII is not responsible for this phenomenon. Enzymatic characterization of PyrKII revealed activity against all NDPs and dNDPs tested, suggesting that it may be capable of generating a broad range of nucleotide triphosphates. Conditional mislocalization of PyrKII resulted in decreased transcript levels within the apicoplast that preceded organelle disruption, suggesting that PyrKII is required for organelle maintenance due to its role in nucleotide triphosphate generation. Introduction *For correspondence: With increasing resistance to current front-line antimalarials, there is a crucial need to find new thera- [email protected] peutic interventions with novel mechanisms of action (Dondorp et al., 2009; Trape, 2001). The api- coplast organelle within the parasite has often been considered as a source of new drug targets Competing interests: The since it is required for blood-stage survival, in addition to possessing evolutionarily distinct biochem- authors declare that no ical pathways that are not present in the human host (Goodman and McFadden, 2013; competing interests exist. -

Catalogue of Protozoan Parasites Recorded in Australia Peter J. O
1 CATALOGUE OF PROTOZOAN PARASITES RECORDED IN AUSTRALIA PETER J. O’DONOGHUE & ROBERT D. ADLARD O’Donoghue, P.J. & Adlard, R.D. 2000 02 29: Catalogue of protozoan parasites recorded in Australia. Memoirs of the Queensland Museum 45(1):1-164. Brisbane. ISSN 0079-8835. Published reports of protozoan species from Australian animals have been compiled into a host- parasite checklist, a parasite-host checklist and a cross-referenced bibliography. Protozoa listed include parasites, commensals and symbionts but free-living species have been excluded. Over 590 protozoan species are listed including amoebae, flagellates, ciliates and ‘sporozoa’ (the latter comprising apicomplexans, microsporans, myxozoans, haplosporidians and paramyxeans). Organisms are recorded in association with some 520 hosts including mammals, marsupials, birds, reptiles, amphibians, fish and invertebrates. Information has been abstracted from over 1,270 scientific publications predating 1999 and all records include taxonomic authorities, synonyms, common names, sites of infection within hosts and geographic locations. Protozoa, parasite checklist, host checklist, bibliography, Australia. Peter J. O’Donoghue, Department of Microbiology and Parasitology, The University of Queensland, St Lucia 4072, Australia; Robert D. Adlard, Protozoa Section, Queensland Museum, PO Box 3300, South Brisbane 4101, Australia; 31 January 2000. CONTENTS the literature for reports relevant to contemporary studies. Such problems could be avoided if all previous HOST-PARASITE CHECKLIST 5 records were consolidated into a single database. Most Mammals 5 researchers currently avail themselves of various Reptiles 21 electronic database and abstracting services but none Amphibians 26 include literature published earlier than 1985 and not all Birds 34 journal titles are covered in their databases. Fish 44 Invertebrates 54 Several catalogues of parasites in Australian PARASITE-HOST CHECKLIST 63 hosts have previously been published. -

Mitosomes in Entamoeba Histolytica Contain a Sulfate Activation Pathway
Mitosomes in Entamoeba histolytica contain a sulfate activation pathway Fumika Mi-ichi, Mohammad Abu Yousuf, Kumiko Nakada-Tsukui, and Tomoyoshi Nozaki1 Department of Parasitology, National Institute of Infectious Diseases, Tokyo 162-8640, Japan Edited by Andrew Roger, Dalhousie University, Halifax, NS, Canada, and accepted by the Editorial Board October 19, 2009 (received for review June 25, 2009) Hydrogenosomes and mitosomes are mitochondrion-related or- and M. balamuthi lack the ISC system, and instead possess the ganelles in anaerobic/microaerophilic eukaryotes with highly re- nitrogen fixation (NIF) system, which is most likely derived from an duced and divergent functions. The full diversity of their content ancestral nitrogen fixing -proteobacterium by lateral gene transfer and function, however, has not been fully determined. To under- (22, 24). Only 5 proteins have been demonstrated in E. histolytica stand the central role of mitosomes in Entamoeba histolytica,a mitosomes: Cpn60 (8–10, 12), Cpn10 (13), mitochondrial Hsp70 parasitic protozoon that causes amoebic dysentery and liver ab- (11, 15), pyridine nucleotide transhydrogenase (PNT) (2, 8), and scesses, we examined the proteomic profile of purified mitosomes. mitochondria carrier family (MCF, ADP/ATP transporter) (14), Using 2 discontinuous Percoll gradient centrifugation and MS and the central role of mitosomes in E. histolytica remains unknown. analysis, we identified 95 putative mitosomal proteins. Immuno- Analysis of the genome of E. histolytica has not revealed any fluorescence assay showed that 3 proteins involved in sulfate additional information regarding the function of mitosomes and activation, ATP sulfurylase, APS kinase, and inorganic pyrophos- thus, a proteomic analysis of mitosomes seems to be the best phatase, as well as sodium/sulfate symporter, involved in sulfate approach to understand its structure and function (1, 2).