The Evolution and Phylogenetic Placement of Invasive Australian
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Materialsupplementary Material
10.1071/BT13149_AC © CSIRO 2013 Australian Journal of Botany 2013, 61(6), 436–445 SUPPLEMENTARY MATERIAL Comparative dating of Acacia: combining fossils and multiple phylogenies to infer ages of clades with poor fossil records Joseph T. MillerA,E, Daniel J. MurphyB, Simon Y. W. HoC, David J. CantrillB and David SeiglerD ACentre for Australian National Biodiversity Research, CSIRO Plant Industry, GPO Box 1600 Canberra, ACT 2601, Australia. BRoyal Botanic Gardens Melbourne, Birdwood Avenue, South Yarra, Vic. 3141, Australia. CSchool of Biological Sciences, Edgeworth David Building, University of Sydney, Sydney, NSW 2006, Australia. DDepartment of Plant Biology, University of Illinois, Urbana, IL 61801, USA. ECorresponding author. Email: [email protected] Table S1 Materials used in the study Taxon Dataset Genbank Acacia abbreviata Maslin 2 3 JF420287 JF420065 JF420395 KC421289 KC796176 JF420499 Acacia adoxa Pedley 2 3 JF420044 AF523076 AF195716 AF195684; AF195703 Acacia ampliceps Maslin 1 KC421930 EU439994 EU811845 Acacia anceps DC. 2 3 JF420244 JF420350 JF419919 JF420130 JF420456 Acacia aneura F.Muell. ex Benth 2 3 JF420259 JF420036 JF420366 JF419935 JF420146 KF048140 Acacia aneura F.Muell. ex Benth. 1 2 3 JF420293 JF420402 KC421323 JQ248740 JF420505 Acacia baeuerlenii Maiden & R.T.Baker 2 3 JF420229 JQ248866 JF420336 JF419909 JF420115 JF420448 Acacia beckleri Tindale 2 3 JF420260 JF420037 JF420367 JF419936 JF420147 JF420473 Acacia cochlearis (Labill.) H.L.Wendl. 2 3 KC283897 KC200719 JQ943314 AF523156 KC284140 KC957934 Acacia cognata Domin 2 3 JF420246 JF420022 JF420352 JF419921 JF420132 JF420458 Acacia cultriformis A.Cunn. ex G.Don 2 3 JF420278 JF420056 JF420387 KC421263 KC796172 JF420494 Acacia cupularis Domin 2 3 JF420247 JF420023 JF420353 JF419922 JF420133 JF420459 Acacia dealbata Link 2 3 JF420269 JF420378 KC421251 KC955787 JF420485 Acacia dealbata Link 2 3 KC283375 KC200761 JQ942686 KC421315 KC284195 Acacia deanei (R.T.Baker) M.B.Welch, Coombs 2 3 JF420294 JF420403 KC421329 KC955795 & McGlynn JF420506 Acacia dempsteri F.Muell. -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

Native Plants Sixth Edition Sixth Edition AUSTRALIAN Native Plants Cultivation, Use in Landscaping and Propagation
AUSTRALIAN NATIVE PLANTS SIXTH EDITION SIXTH EDITION AUSTRALIAN NATIVE PLANTS Cultivation, Use in Landscaping and Propagation John W. Wrigley Murray Fagg Sixth Edition published in Australia in 2013 by ACKNOWLEDGEMENTS Reed New Holland an imprint of New Holland Publishers (Australia) Pty Ltd Sydney • Auckland • London • Cape Town Many people have helped us since 1977 when we began writing the first edition of Garfield House 86–88 Edgware Road London W2 2EA United Kingdom Australian Native Plants. Some of these folk have regrettably passed on, others have moved 1/66 Gibbes Street Chatswood NSW 2067 Australia to different areas. We endeavour here to acknowledge their assistance, without which the 218 Lake Road Northcote Auckland New Zealand Wembley Square First Floor Solan Road Gardens Cape Town 8001 South Africa various editions of this book would not have been as useful to so many gardeners and lovers of Australian plants. www.newhollandpublishers.com To the following people, our sincere thanks: Steve Adams, Ralph Bailey, Natalie Barnett, www.newholland.com.au Tony Bean, Lloyd Bird, John Birks, Mr and Mrs Blacklock, Don Blaxell, Jim Bourner, John Copyright © 2013 in text: John Wrigley Briggs, Colin Broadfoot, Dot Brown, the late George Brown, Ray Brown, Leslie Conway, Copyright © 2013 in map: Ian Faulkner Copyright © 2013 in photographs and illustrations: Murray Fagg Russell and Sharon Costin, Kirsten Cowley, Lyn Craven (Petraeomyrtus punicea photograph) Copyright © 2013 New Holland Publishers (Australia) Pty Ltd Richard Cummings, Bert -

503 Flora V7 2.Doc 3
Browse LNG Precinct ©WOODSIDE Browse Liquefied Natural Gas Precinct Strategic Assessment Report (Draft for Public Review) December 2010 Appendix C-18 A Vegetation and Flora Survey of James Price Point: Wet Season 2009 A Vegetation and Flora Survey of James Price Point: Wet Season 2009 Prepared for Department of State Development December 2009 A Vegetation and Flora Survey of James Price Point: Wet Season 2009 © Biota Environmental Sciences Pty Ltd 2009 ABN 49 092 687 119 Level 1, 228 Carr Place Leederville Western Australia 6007 Ph: (08) 9328 1900 Fax: (08) 9328 6138 Project No.: 503 Prepared by: P. Chukowry, M. Maier Checked by: G. Humphreys Approved for Issue: M. Maier This document has been prepared to the requirements of the client identified on the cover page and no representation is made to any third party. It may be cited for the purposes of scientific research or other fair use, but it may not be reproduced or distributed to any third party by any physical or electronic means without the express permission of the client for whom it was prepared or Biota Environmental Sciences Pty Ltd. This report has been designed for double-sided printing. Hard copies supplied by Biota are printed on recycled paper. Cube:Current:503 (Kimberley Hub Wet Season):Doc:Flora:503 flora v7_2.doc 3 A Vegetation and Flora Survey of James Price Point: Wet Season 2009 4 Cube:Current:503 (Kimberley Hub Wet Season):Doc:Flora:503 flora v7_2.doc Biota A Vegetation and Flora Survey of James Price Point: Wet Season 2009 A Vegetation and Flora Survey of James Price -
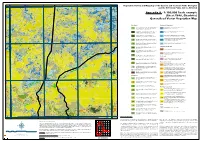
Appendix 9 - 1:100,000 Scale Example (Sheet 5648, Charlotte) Generalised Vector Vegetation Map
133°30'E 133°40'E 133°50'E 134°E 24°30'S Vegetation Survey and Mapping of the Eastern and Southern Finke Bioregion 24°30'S and the NT Stony Plains Inliers, NT & SA Appendix 9 - 1:100,000 Scale example (Sheet 5648, Charlotte) Generalised Vector Vegetation Map Woodland Chenopod Shrubland Acacia aneura ( Mulga) Low Open Woodland TO Tall Open Shrubland of Atriplex nummularia (Old man saltbush) Low Sparse Chenopod 1 Acacia estrophiolata (Ironwood) on clay loam plains and red earth 4 shrubland over Low Sparse Tussock grasses. soils+/- Atriplex vesicaria and Eragrostis eriopoda . Acacia georginae / Acacia cambagei ( Gidgee) Low Woodland to Tall Atriplex vesicaria (Pop saltbush) Low Open Chenopod Shrubland.+/- 2 Shrubland.+/- Eucalyptus coolabah subsp. Arida , Codonocarpus 5 Maireana astrotricha over tussock grasses. cotinifolius , Eulalia aurea, Eriachne ovata and Atriplex vesicaria . Eucalyptus coolabah subsp. arida (Coolabah) Woodland. +/- Maireana aphylla (Cottonbush) Low Sparse Chenopod Shrubland. +/- 12 Muehlenbeckia florulenta , Acacia aneura , Senna artemisioides ssp. 8 Fimbristylis dichotoma , Dactyloctenium radulans and Eragrostis dielsii. Filifolia , Marsilea sp ., Cynodon dactylon , and Cenchrus ciliaris . Maireana astrotricha (Low bluebush) Low Sparse Chenopod Shrubland Eucalyptus camaldulensis var. obtusa (River red gum) Woodland.+/- TO Sparse shrubland of Senna artemisioides n. coriacea and 13 Eucalyptus coolabah subsp. arida , Cynodon dactylon , Eulalia aurea and 9 Eremophila duttonii (Harlequin fuchsia bush). Cyperus gymnocaulos . 24°40'S Hakea leucoptera subsp. leucoptera (Needlewood) Open Woodland. +- Sclerolaena (mixed) Low Sparse Chenopod Shrubland.+/- Enneapogon 24°40'S 14 Eremophila sturtii , Senna artemisioides ssp. filifolia , Hakea leucoptera 15 avenaceus Aristida contorta , Sporobolus actinocladus . subsp. leucoptera and Triodia basedowii . Acacia calcicola (Northern Myall) Sparse Woodland +/- Eremophila Samphire Shrubland 23 duttonii , Acacia calcicola , Atriplex vesicaria , Maireana georgei and mixed short grasses. -

Evolution of Angiosperm Pollen. 7. Nitrogen-Fixing Clade1
Evolution of Angiosperm Pollen. 7. Nitrogen-Fixing Clade1 Authors: Jiang, Wei, He, Hua-Jie, Lu, Lu, Burgess, Kevin S., Wang, Hong, et. al. Source: Annals of the Missouri Botanical Garden, 104(2) : 171-229 Published By: Missouri Botanical Garden Press URL: https://doi.org/10.3417/2019337 BioOne Complete (complete.BioOne.org) is a full-text database of 200 subscribed and open-access titles in the biological, ecological, and environmental sciences published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Complete website, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/terms-of-use. Usage of BioOne Complete content is strictly limited to personal, educational, and non - commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Downloaded From: https://bioone.org/journals/Annals-of-the-Missouri-Botanical-Garden on 01 Apr 2020 Terms of Use: https://bioone.org/terms-of-use Access provided by Kunming Institute of Botany, CAS Volume 104 Annals Number 2 of the R 2019 Missouri Botanical Garden EVOLUTION OF ANGIOSPERM Wei Jiang,2,3,7 Hua-Jie He,4,7 Lu Lu,2,5 POLLEN. 7. NITROGEN-FIXING Kevin S. Burgess,6 Hong Wang,2* and 2,4 CLADE1 De-Zhu Li * ABSTRACT Nitrogen-fixing symbiosis in root nodules is known in only 10 families, which are distributed among a clade of four orders and delimited as the nitrogen-fixing clade. -

Indigenous Plants of Bendigo
Produced by Indigenous Plants of Bendigo Indigenous Plants of Bendigo PMS 1807 RED PMS 432 GREY PMS 142 GOLD A Gardener’s Guide to Growing and Protecting Local Plants 3rd Edition 9 © Copyright City of Greater Bendigo and Bendigo Native Plant Group Inc. This work is Copyright. Apart from any use permitted under the Copyright Act 1968, no part may be reproduced by any process without prior written permission from the City of Greater Bendigo. First Published 2004 Second Edition 2007 Third Edition 2013 Printed by Bendigo Modern Press: www.bmp.com.au This book is also available on the City of Greater Bendigo website: www.bendigo.vic.gov.au Printed on 100% recycled paper. Disclaimer “The information contained in this publication is of a general nature only. This publication is not intended to provide a definitive analysis, or discussion, on each issue canvassed. While the Committee/Council believes the information contained herein is correct, it does not accept any liability whatsoever/howsoever arising from reliance on this publication. Therefore, readers should make their own enquiries, and conduct their own investigations, concerning every issue canvassed herein.” Front cover - Clockwise from centre top: Bendigo Wax-flower (Pam Sheean), Hoary Sunray (Marilyn Sprague), Red Ironbark (Pam Sheean), Green Mallee (Anthony Sheean), Whirrakee Wattle (Anthony Sheean). Table of contents Acknowledgements ...............................................2 Foreword..........................................................3 Introduction.......................................................4 -

Table of Contents Below) with Family Name Provided
1 Australian Plants Society Plant Table Profiles – Sutherland Group (updated August 2021) Below is a progressive list of all cultivated plants from members’ gardens and Joseph Banks Native Plants Reserve that have made an appearance on the Plant Table at Sutherland Group meetings. Links to websites are provided for the plants so that further research can be done. Plants are grouped in the categories of: Trees and large shrubs (woody plants generally taller than 4 m) Medium to small shrubs (woody plants from 0.1 to 4 m) Ground covers or ground-dwelling (Grasses, orchids, herbaceous and soft-wooded plants, ferns etc), as well as epiphytes (eg: Platycerium) Vines and scramblers Plants are in alphabetical order by botanic names within plants categories (see table of contents below) with family name provided. Common names are included where there is a known common name for the plant: Table of Contents Trees and Large shrubs........................................................................................................................... 2 Medium to small shrubs ...................................................................................................................... 23 Groundcovers and other ground‐dwelling plants as well as epiphytes. ............................................ 64 Vines and Scramblers ........................................................................................................................... 86 Sutherland Group http://sutherland.austplants.com.au 2 Trees and Large shrubs Acacia decurrens -

A Taxonomic Revision of Acacia Vernicifluaand A. Leprosa
A taxonomic revision of Acacia verniciflua and A. leprosa (Leguminosae: Mimosoideae) in Australia Bruce R. Maslin 1 and Daniel J. Murphy 2 1 Western Australian Herbarium, Department of Environment and Conservation, Locked Bag 104, Bentley Delivery Centre, Western Australia 6983, Australia; e-mail: [email protected] 2 National Herbarium of Victoria, Royal Botanic Gardens Melbourne, Birdwood Avenue, South Yarra, Victoria 3141, Australia; e-mail: [email protected] Introduction Abstract Acacia verniciflua (Varnish Wattle) and A. leprosa (Cinnamon Wattle) A revision of a taxonomically complex occur in temperate areas of eastern and southern Australia and as group of species allied to, and including, Acacia leprosa Sieber ex DC. hitherto defined were regarded as highly polymorphic species in need of and A. verniciflua A.Cunn. is presented. critical revision (Maslin 2001). The conventional separation between the These species predominate in species was the number of longitudinal nerves on their phyllodes, one in temperate regions of eastern Australia. A. leprosa and two in A. verniciflua, but as correctly noted by Court (1972, Two new species are described, p. 219), the importance of this character has been over-emphasised. A. rostriformis Maslin & D.J.Murphy and A. stictophylla Court ex Maslin During the nineteenth century a number of taxa were described that & D.J.Murphy and one, A. exudans were referable to the A. verniciflua– A. leprosa group, however, none Lindl., is resurrected. Acacia leprosa is of these names was ever taken up, presumably because of difficulties treated as a highly polymorphic species in defining the taxa and uncertainties concerning the application of comprising five varieties, four of which the names (some of which were based on plants cultivated in Europe are described as new, namely, A. -

Flora Survey on Hiltaba Station and Gawler Ranges National Park
Flora Survey on Hiltaba Station and Gawler Ranges National Park Hiltaba Pastoral Lease and Gawler Ranges National Park, South Australia Survey conducted: 12 to 22 Nov 2012 Report submitted: 22 May 2013 P.J. Lang, J. Kellermann, G.H. Bell & H.B. Cross with contributions from C.J. Brodie, H.P. Vonow & M. Waycott SA Department of Environment, Water and Natural Resources Vascular plants, macrofungi, lichens, and bryophytes Bush Blitz – Flora Survey on Hiltaba Station and Gawler Ranges NP, November 2012 Report submitted to Bush Blitz, Australian Biological Resources Study: 22 May 2013. Published online on http://data.environment.sa.gov.au/: 25 Nov. 2016. ISBN 978-1-922027-49-8 (pdf) © Department of Environment, Water and Natural Resouces, South Australia, 2013. With the exception of the Piping Shrike emblem, images, and other material or devices protected by a trademark and subject to review by the Government of South Australia at all times, this report is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. All other rights are reserved. This report should be cited as: Lang, P.J.1, Kellermann, J.1, 2, Bell, G.H.1 & Cross, H.B.1, 2, 3 (2013). Flora survey on Hiltaba Station and Gawler Ranges National Park: vascular plants, macrofungi, lichens, and bryophytes. Report for Bush Blitz, Australian Biological Resources Study, Canberra. (Department of Environment, Water and Natural Resources, South Australia: Adelaide). Authors’ addresses: 1State Herbarium of South Australia, Department of Environment, Water and Natural Resources (DEWNR), GPO Box 1047, Adelaide, SA 5001, Australia. -

ACT, Australian Capital Territory
Biodiversity Summary for NRM Regions Species List What is the summary for and where does it come from? This list has been produced by the Department of Sustainability, Environment, Water, Population and Communities (SEWPC) for the Natural Resource Management Spatial Information System. The list was produced using the AustralianAustralian Natural Natural Heritage Heritage Assessment Assessment Tool Tool (ANHAT), which analyses data from a range of plant and animal surveys and collections from across Australia to automatically generate a report for each NRM region. Data sources (Appendix 2) include national and state herbaria, museums, state governments, CSIRO, Birds Australia and a range of surveys conducted by or for DEWHA. For each family of plant and animal covered by ANHAT (Appendix 1), this document gives the number of species in the country and how many of them are found in the region. It also identifies species listed as Vulnerable, Critically Endangered, Endangered or Conservation Dependent under the EPBC Act. A biodiversity summary for this region is also available. For more information please see: www.environment.gov.au/heritage/anhat/index.html Limitations • ANHAT currently contains information on the distribution of over 30,000 Australian taxa. This includes all mammals, birds, reptiles, frogs and fish, 137 families of vascular plants (over 15,000 species) and a range of invertebrate groups. Groups notnot yet yet covered covered in inANHAT ANHAT are notnot included included in in the the list. list. • The data used come from authoritative sources, but they are not perfect. All species names have been confirmed as valid species names, but it is not possible to confirm all species locations. -

MVG 16 Acacia Shrublands DRAFT
MVG 16 - ACACIA SHRUBLANDS Acacia hillii, Tanami Desert, NT (Photo: D. Keith) Overview The overstorey of MVG 16 is dominated by multi-stemmed acacia shrubs. The most widespread species is Acacia aneura (mulga). Mulga vegetation takes on a variety of structural expressions and is consequently classified partly within MVG 16 where the overstorey is dominated by multi-stemmed shrubs, partly within MVG 6 in accordance with the Kyoto Protocol definition of forest cover in Australia (trees > 2 m tall and crown cover > 20%, foliage projective cover > 10%); and partly within MVG 13 where the woody dominants are predominantly single-stemmed, but with crown cover less than 20%. Occurs where annual rainfall is below 250mm in southern Australia and below 350mm in northern Australia (Hodgkinson 2002; Foulkes et al. 2014). Species composition varies along rainfall gradients, with substrate and rainfall seasonality (Beadle 1981; Johnson and Burrows 1994). Transitions into MVG 13 Acacia woodlands with higher rainfall and varying soil types. Is most commonly found on red earth soils (Hodgkinson 2002). Facts and figures Major Vegetation Group MVG 16 - Acacia Shrublands Major Vegetation Subgroups 20. Stony mulga woodlands and shrublands NSW, (number of NVIS descriptions) NT, QLD, SA, WA 23. Sandplain Acacia woodlands and shrublands NSW, NT, QLD, SA, WA Typical NVIS structural formations Shrubland (tall, mid,) Open shrubland (tall, mid,) Sparse shrubland (tall, mid,) Number of IBRA regions 53 Most extensive in IBRA region Est. pre-1750 and present : Great Victoria Desert (WA and SA) Estimated pre-1750 extent (km2) 865 845 Present extent (km2) 851 274 Area protected (km2) 85 444 Acacia ligulata (sandhill wattle), SA (Photo: M.