Introduction, Literature Review and Thesis Objectives 1.1 Introduction
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Radar.Brookes.Ac.Uk/Radar/Items/C664d3c7-1663-4D34-Aed5-E3e070598081/1
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Oxford Brookes University: RADAR RADAR Research Archive and Digital Asset Repository Pozzi, L, Nekaris, K, Perkin, A, Bearder, S, Pimley, E, Schulze, H, Streicher, U, Nadler, T, Kitchener, A, Zischler, H, Zinner, D and Roos, C Remarkable Ancient Divergences Amongst Neglected Lorisiform Primates Pozzi, L, Nekaris, K, Perkin, A, Bearder, S, Pimley, E, Schulze, H, Streicher, U, Nadler, T, Kitchener, A, Zischler, H, Zinner, D and Roos, C (2015) Remarkable Ancient Divergences Amongst Neglected Lorisiform Primates. Zoological Journal of the Linnean Society, 175 (3). pp. 661-674. doi: 10.1111/zoj.12286 This version is available: https://radar.brookes.ac.uk/radar/items/c664d3c7-1663-4d34-aed5-e3e070598081/1/ Available on RADAR: August 2016 Copyright © and Moral Rights are retained by the author(s) and/ or other copyright owners. A copy can be downloaded for personal non-commercial research or study, without prior permission or charge. This item cannot be reproduced or quoted extensively from without first obtaining permission in writing from the copyright holder(s). The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the copyright holders. This document is the published version of the journal article. WWW.BROOKES.AC.UK/GO/RADAR bs_bs_banner Zoological Journal of the Linnean Society, 2015, 175, 661–674. With 3 figures Remarkable ancient divergences amongst neglected lorisiform primates LUCA -

Achatina Fulica and Archachatina Marginata Was Sampled in the Littoral, Center and West Regions of Cameroon
Central Journal of Veterinary Medicine and Research Research Article *Corresponding author Prisca Meffowoet, Faculty of Agronomy and Agricul- tural Sciences, University of Dschang, Dschang, Tel: Infestation rate of African 699366311/676879291; Email: [email protected] Submitted: 25 March 2020 giant snails (Achatina fulica Accepted: 02 April 2020 Published: 06 April 2020 ISSN: 2379-948X and Archachatina marginata) by Copyright © 2020 Prisca MC, et al. parasites during the rainy season OPEN ACCESS Keywords • Cameroon in three localities of Cameroon • African giant snails • Parasites Meffowoet CP1*, Kouam KM1, Kana JR2, and Tchakounte FM2 1Animal Physiology and Health Research Unit, University of Dschang, Cameroon 2Animal Nutrition and Production Research Unit, University of Dschang, Cameroon Abstract This study was designed during the rainy season in order to identify the parasites likely to infest edible snails. 360 Achatina fulica and Archachatina marginata was sampled in the Littoral, Center and West regions of Cameroon. After macroscopic observation of snails, the hepatopancreas, digestive tract, sex organs, slime and haemolymph were isolated. These samples were examined using the flotation techniques and direct rubbing. Of the 360 snails sampled, 213 (59.3%) were infested, that is 147 (82.1%) for A. marginata and 66 (36.7%) for A. fulica respectively. The highest infestation rate was recorded on protozoans (41.4%) followed by nematode (24.7%). The most represented parasites were Trichodina achatinae (23.9%) and Strongyloides stercoralis (16.1%); while the least represented were cyst of Balantidium coli (8.1%), Enteromonas sp. (8.1%), cyst of Isospora sp. (7.8%), larva of Protostrongylus sp. (7.5%), cyst of Cryptosporidium sp. -

The Gastropod Shell Has Been Co-Opted to Kill Parasitic Nematodes
www.nature.com/scientificreports OPEN The gastropod shell has been co- opted to kill parasitic nematodes R. Rae Exoskeletons have evolved 18 times independently over 550 MYA and are essential for the success of Received: 23 March 2017 the Gastropoda. The gastropod shell shows a vast array of different sizes, shapes and structures, and Accepted: 18 May 2017 is made of conchiolin and calcium carbonate, which provides protection from predators and extreme Published: xx xx xxxx environmental conditions. Here, I report that the gastropod shell has another function and has been co-opted as a defense system to encase and kill parasitic nematodes. Upon infection, cells on the inner layer of the shell adhere to the nematode cuticle, swarm over its body and fuse it to the inside of the shell. Shells of wild Cepaea nemoralis, C. hortensis and Cornu aspersum from around the U.K. are heavily infected with several nematode species including Caenorhabditis elegans. By examining conchology collections I show that nematodes are permanently fixed in shells for hundreds of years and that nematode encapsulation is a pleisomorphic trait, prevalent in both the achatinoid and non-achatinoid clades of the Stylommatophora (and slugs and shelled slugs), which diverged 90–130 MYA. Taken together, these results show that the shell also evolved to kill parasitic nematodes and this is the only example of an exoskeleton that has been co-opted as an immune system. The evolution of the shell has aided in the success of the Gastropoda, which are composed of 65–80,000 spe- cies that have colonised terrestrial and marine environments over 400MY1, 2. -

Infant Development in the Slender Loris (Loris Lydekkerianus Lydekkerianus)
RESEARCH ARTICLES Infant development in the slender loris (Loris lydekkerianus lydekkerianus) Sindhu Radhakrishna1,* and Mewa Singh Department of Psychology, University of Mysore, Mysore 570 006, India 1Present address: National Institute of Advanced Studies, Indian Institute of Science Campus, Bangalore 560 012, India lorises and bushbabies)1, the lorids (angwantibos, pottos In this article we present data on infant development in wild slender loris, a nocturnal primate species. The and lorises) typically have lower developmental and repro- ductive rates in comparison to similar-sized galagos (bush- behavioural ecology of the grey slender loris Loris lyde- 2–4 kkerianus lydekkerianus, a nocturnal strepsirrhine, babies) . Litter size in lorids and galagos appears to be was studied for 21 months (October 1997–June 1999) linked to the stability of the habitat more than body size: in a scrub jungle in Dindigul, south India. A total of species occupying harsher environments tend to have 22,834 scans were collected during 2656 h of observa- higher reproductive rates3,5. Strepsirrhine neonates are tion on identified and unidentified lorises using in- carried for short periods following birth and mode of stantaneous point and ad libitum sampling methods. infant transport can be oral (Galagos bushbabies, Cheiro- Developmental schedules were observed for twelve in- galeus dwarf lemurs, Microcebus mouse lemurs) or the dividuals born during the course of the study period. abdominal fur of the mother (Loris slender loris)3,6,7. In- A greater number of twin births were observed than fant parking is typical of the Loriformes, wherein infants singleton births and more isosexuals than heterosexu- als. -

Pest Alert: Giant African Snails
Pest Alert Giant African Snails “Giant African snail” is the Services. This action resulted common name used to describe in the State’s second giant several foreign snail species African snail detection. The that could become serious U.S. Department of Agriculture agricultural pests in the United (USDA) and Florida have States. The most important giant invested several million dollars African snail is Lissachatina since this second detection, fulica (formerly Achatina fulica). and work continues today to Figure 1. A mature Lissachatina fulica eradicate this pest. maneuvers in its environment. The Giant African Snail Description/Life Cycle Scientists consider L. fulica to be one of the most damaging Reaching almost 8 inches land snails in the world. It is (20 centimeters) in length and known to feed on at least 5 inches (13 centimeters) in 500 different types of plants, diameter, L. fulica is one of the including peanuts, beans, peas, world’s largest land snails— cucumbers, and melons. If about the size of an average fruits and vegetables are not adult fist. When fully grown, available, they will eat a wide its shell consists of seven to variety of ornamental plants, tree nine whorls, with a long and bark, and even paint and stucco greatly swollen body whorl. The Figure 2. A penny is used to show the size of on houses. giant African snail eggs. brownish shell with darker brown lengthwise stripes covers at L. fulica is established least half the length of the snail. throughout the Indo-Pacific Florida and the Giant basin, from east Africa to African Snail Each snail contains both Hawaii and Guam, including female and male reproductive the Southern Asian region. -

Biosecurity Risk Assessment
An Invasive Risk Assessment Framework for New Animal and Plant-based Production Industries RIRDC Publication No. 11/141 RIRDCInnovation for rural Australia An Invasive Risk Assessment Framework for New Animal and Plant-based Production Industries by Dr Robert C Keogh February 2012 RIRDC Publication No. 11/141 RIRDC Project No. PRJ-007347 © 2012 Rural Industries Research and Development Corporation. All rights reserved. ISBN 978-1-74254-320-8 ISSN 1440-6845 An Invasive Risk Assessment Framework for New Animal and Plant-based Production Industries Publication No. 11/141 Project No. PRJ-007347 The information contained in this publication is intended for general use to assist public knowledge and discussion and to help improve the development of sustainable regions. You must not rely on any information contained in this publication without taking specialist advice relevant to your particular circumstances. While reasonable care has been taken in preparing this publication to ensure that information is true and correct, the Commonwealth of Australia gives no assurance as to the accuracy of any information in this publication. The Commonwealth of Australia, the Rural Industries Research and Development Corporation (RIRDC), the authors or contributors expressly disclaim, to the maximum extent permitted by law, all responsibility and liability to any person, arising directly or indirectly from any act or omission, or for any consequences of any such act or omission, made in reliance on the contents of this publication, whether or not caused by any negligence on the part of the Commonwealth of Australia, RIRDC, the authors or contributors. The Commonwealth of Australia does not necessarily endorse the views in this publication. -
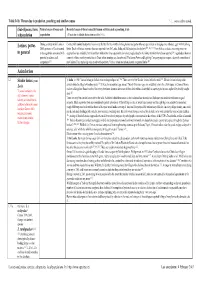
Table 14 B: Threat Due to Predation, Poaching and Similar Causes 1, 2,
Table 14 b: Threat due to predation, poaching and similar causes 1, 2, ... : source, author quoted. (Sub-)Species, form, Natural causes of losses such Recorded causes of threat caused by human activities such as poaching, trade subpopulation as predation (Threat due to habitat destruction see table 14 c) Lorises, pottos During a survey in Sri Lanka in Contact with uninsulated power lines is may be fatal for the slowly climbing lorises and pottos who are specialized on bridging over substrate gaps with their long 2001, presence of Loris seemed limbs. Death of lorises on power lines are reported from Sri Lanka, India and Malaysia (see also below) 207, 66, 211. From Africa, no data concerning pottos or in general to be negatively associated with angwantibos are available, but the problem with power lines apparently also exists, regular deaths of colobus monkeys have been reported 208, regarded as the main potential predators and cause of colobus monkey mortalities in Diani; other monkeys are also affected. The Kenya Power and Lighting Company engineers spent a day with coworkers of competitors 211. the Colobus Trust exploring ways to solve the problem. Victims of electrocution showed amputated limbs 209. Asian lorises L I Slender lorises, genus In India, in 1981 "lorises belong to India's most endangered species" 116. There are very few Slender lorises left in the wild 121. Slender lorises belong to the Loris animals found in illegal ownership (pets) 110. It was also a popular cage animal. Twenty-five years ago, one could buy a loris for a few rupees in Chennai Moore market or Bangalore Russel market. -

Giant African Snail Cooperative Eradication Program
United States Department of Agriculture Giant African Snail Marketing and Regulatory Cooperative Programs Animal and Eradication Program Plant Health Inspection Service Miami-Dade County, Florida Environmental Assessment, October 2011 Giant African Snail Cooperative Eradication Program Environmental Assessment October 2011 Agency Contact: Andrea Simao USDA–APHIS–PPQ Emergency and Domestic Programs 4700 River Road, Unit 26 Riverdale, MD 20737 __________________________________________________________ The U.S. Department of Agriculture (USDA) prohibits discrimination in all its programs and activities on the basis of race, color, national origin, sex, religion, age, disability, political beliefs, sexual orientation, or marital or family status. (Not all prohibited bases apply to all programs.) Persons with disabilities who require alternative means for communication of program information (Braille, large print, audiotape, etc.) should contact USDA’S TARGET Center at (202) 720–2600 (voice and TDD). To file a complaint of discrimination, write USDA, Director, Office of Civil Rights, Room 326–W, Whitten Building, 1400 Independence Avenue, SW, Washington, DC 20250–9410 or call (202) 720–5964 (voice and TDD). USDA is an equal opportunity provider and employer. __________________________________________________________ Mention of companies or commercial products in this report does not imply recommendation or endorsement by the U.S. Department of Agriculture over others not mentioned. USDA neither guarantees nor warrants the standard of any product -

Primate Conservation
Primate Conservation Global evidence for the effects of interventions Jessica Junker, Hjalmar S. Kühl, Lisa Orth, Rebecca K. Smith, Silviu O. Petrovan and William J. Sutherland Synopses of Conservation Evidence ii © 2017 William J. Sutherland This work is licensed under a Creative Commons Attribution 4.0 International license (CC BY 4.0). This license allows you to share, copy, distribute and transmit the work; to adapt the work and to make commercial use of the work providing attribution is made to the authors (but not in any way that suggests that they endorse you or your use of the work). Attribution should include the following information: Junker, J., Kühl, H.S., Orth, L., Smith, R.K., Petrovan, S.O. and Sutherland, W.J. (2017) Primate conservation: Global evidence for the effects of interventions. University of Cambridge, UK Further details about CC BY licenses are available at https://creativecommons.org/licenses/by/4.0/ Cover image: Martha Robbins/MPI-EVAN Bwindi Impenetrable National Park, Uganda Digital material and resources associated with this synopsis are available at https://www.conservationevidence.com/ iii Contents About this book ............................................................................................................................. xiii 1. Threat: Residential and commercial development ............................ 1 Key messages ........................................................................................................................................ 1 1.1. Remove and relocate ‘problem’ -

Life After Logging: Reconciling Wildlife Conservation and Production Forestry in Indonesian Borneo
Life after logging Reconciling wildlife conservation and production forestry in Indonesian Borneo Erik Meijaard • Douglas Sheil • Robert Nasi • David Augeri • Barry Rosenbaum Djoko Iskandar • Titiek Setyawati • Martjan Lammertink • Ike Rachmatika • Anna Wong Tonny Soehartono • Scott Stanley • Timothy O’Brien Foreword by Professor Jeffrey A. Sayer Life after logging: Reconciling wildlife conservation and production forestry in Indonesian Borneo Life after logging: Reconciling wildlife conservation and production forestry in Indonesian Borneo Erik Meijaard Douglas Sheil Robert Nasi David Augeri Barry Rosenbaum Djoko Iskandar Titiek Setyawati Martjan Lammertink Ike Rachmatika Anna Wong Tonny Soehartono Scott Stanley Timothy O’Brien With further contributions from Robert Inger, Muchamad Indrawan, Kuswata Kartawinata, Bas van Balen, Gabriella Fredriksson, Rona Dennis, Stephan Wulffraat, Will Duckworth and Tigga Kingston © 2005 by CIFOR and UNESCO All rights reserved. Published in 2005 Printed in Indonesia Printer, Jakarta Design and layout by Catur Wahyu and Gideon Suharyanto Cover photos (from left to right): Large mature trees found in primary forest provide various key habitat functions important for wildlife. (Photo by Herwasono Soedjito) An orphaned Bornean Gibbon (Hylobates muelleri), one of the victims of poor-logging and illegal hunting. (Photo by Kimabajo) Roads lead to various impacts such as the fragmentation of forest cover and the siltation of stream— other impacts are associated with improved accessibility for people. (Photo by Douglas Sheil) This book has been published with fi nancial support from UNESCO, ITTO, and SwedBio. The authors are responsible for the choice and presentation of the facts contained in this book and for the opinions expressed therein, which are not necessarily those of CIFOR, UNESCO, ITTO, and SwedBio and do not commit these organisations. -

Foraging Behaviour of the Slender Loris (Loris Lydekkerianus Lydekkerianus): Implications for Theories of Primate Origins
Journal of Human Evolution 49 (2005) 289e300 Foraging behaviour of the slender loris (Loris lydekkerianus lydekkerianus): implications for theories of primate origins K.A.I. Nekaris* Department of Anthropology, Washington University, Campus Box 1114, One Brookings Drive, St. Louis, MO 63110, USA Received 3 May 2004; accepted 18 April 2005 Abstract Members of the Order Primates are characterised by a wide overlap of visual fields or optic convergence. It has been proposed that exploitation of either insects or angiosperm products in the terminal branches of trees, and the corresponding complex, three-dimensional environment associated with these foraging strategies, account for visual convergence. Although slender lorises (Loris sp.) are the most visually convergent of all the primates, very little is known about their feeding ecology. This study, carried out over 10 ½ months in South India, examines the feeding behaviour of L. lydekkerianus lydekkerianus in relation to hypotheses regarding visual predation of insects. Of 1238 feeding observations, 96% were of animal prey. Lorises showed an equal and overwhelming preference for terminal and middle branch feeding, using the undergrowth and trunk rarely. The type of prey caught on terminal branches (Lepidoptera, Odonata, Homoptera) differed significantly from those caught on middle branches (Hymenoptera, Coleoptera). A two-handed catch accompanied by bipedal postures was used almost exclusively on terminal branches where mobile prey was caught, whereas the more common capture technique of one-handed grab was used more often on sturdy middle branches to obtain slow moving prey. Although prey was detected with senses other than vision, vision was the key sense used upon the final strike. -

The Giant African Land Snail (Lissachatina Fulica)
The Giant African Land Snail (Lissachatina fulica) Photo: Yuri Yashin, achatina.ru, www.bugwood.org, #1265024 The Giant African Land Snail Adult • Invasive mollusk • Established in Florida and under eradication in Miami-Dade County. • Live snails in this genus are specifically prohibited from both interstate movement and importation into the U.S. Photo: Yuri Yashin, achatina.ru, www.bugwood.org, #1265024 Distribution in Florida • Populations in Miami- Dade Co. • Active eradication effort • Report suspected sightings to: No sampling (888) 397-1517 DetectedNo sampling Sampled but not found Found, but not established Map courtesy of Florida Department of Agriculture and Consumer Services Potential U.S. Range Potential Distribution Map based on http://www.aphis.usda.gov/import_export/plants/manuals/emergency/downloads/nprg_gas.pdf Susceptible Plants Photos: Top Row - Charles T. Bryson, USDA Agricultural Research Service, www.bugwood.org, #1116132; #1197011; Forest & Kim Starr, Starr Environmental, www.bugwood.org, #5420178; Bottom Row - Howard F. Schwartz, Colorado State University, www.bugwood.org, #5365883; #5363704; - M.E. Bartolo, www.bugwood.org, #5359190 Identification: Eggs Photos: Top left – Lyle Buss, Department of Entomology and Nematology, University of Florida; Bottom left - David Robinson, USDA-APHIS-PPQ; Right - Yuri Yashin, achatina.ru, www.bugwood.org, #1265029 Identification: Juveniles “Truncated Columella” “Outer lip” Photos: Left and middle - Lyle Buss, Department of Entomology and Nematology, University of Florida Right