Genetic Structure of Remnant Quercus Serrata Populations at The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
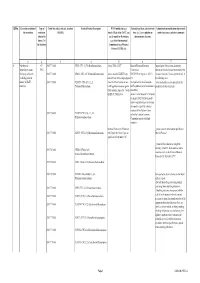
Qrno. 1 2 3 4 5 6 7 1 CP 2903 77 100 0 Cfcl3
QRNo. General description of Type of Tariff line code(s) affected, based on Detailed Product Description WTO Justification (e.g. National legal basis and entry into Administration, modification of previously the restriction restriction HS(2012) Article XX(g) of the GATT, etc.) force (i.e. Law, regulation or notified measures, and other comments (Symbol in and Grounds for Restriction, administrative decision) Annex 2 of e.g., Other International the Decision) Commitments (e.g. Montreal Protocol, CITES, etc) 12 3 4 5 6 7 1 Prohibition to CP 2903 77 100 0 CFCl3 (CFC-11) Trichlorofluoromethane Article XX(h) GATT Board of Eurasian Economic Import/export of these ozone destroying import/export ozone CP-X Commission substances from/to the customs territory of the destroying substances 2903 77 200 0 CF2Cl2 (CFC-12) Dichlorodifluoromethane Article 46 of the EAEU Treaty DECISION on August 16, 2012 N Eurasian Economic Union is permitted only in (excluding goods in dated 29 may 2014 and paragraphs 134 the following cases: transit) (all EAEU 2903 77 300 0 C2F3Cl3 (CFC-113) 1,1,2- 4 and 37 of the Protocol on non- On legal acts in the field of non- _to be used solely as a raw material for the countries) Trichlorotrifluoroethane tariff regulation measures against tariff regulation (as last amended at 2 production of other chemicals; third countries Annex No. 7 to the June 2016) EAEU of 29 May 2014 Annex 1 to the Decision N 134 dated 16 August 2012 Unit list of goods subject to prohibitions or restrictions on import or export by countries- members of the -

Concern About Sudden Oak Deatn Grows
The Newsletter of the International Oak Society, Volume 8, No. 2, july 2004 Concern About Sudden Oak Deatn Grows As reported in several issues of California, Oregon, Ohio, North complete and the nursery is found this newsletter, and in Proceed Carolina, and Georgia. The other to be free from the pathogen, all ings articles from the last two In San Diego County nursery con out-of-state shipments of host ternational Oak Society Symposia, ducts much of their sales through nursery stock and associated ar Sudden Oak Death or SOD is a new mail orders. This news has sent ticles, as well as plants within the disease affecting some species of shock waves through the nursery same genus as any host or asso oaks in California. The agent re and forest industries since it was ciated article, and any plant lo sponsible for this disease is feared that infected plants had cated within 10 meters of a host Phytophthora ramorum, a fungus been shipped throughout the or associated article, must remain like water mold that can girdle United States. Trace-back and on hold. For a complete list of mature trees and consequently kill trace-forward surveys were there hosts and associated plants, as them. To date this disease has fore conducted to determine well as the complete text of the been reported on four of where infected plants originated order, go to www.aphis.usda.gov/ California's 20 species of native and where they were shipped once ppq/ispm/sod/index.html. For in oaks - all members of the black they left each of the nurseries. -

New York City Approved Street Trees
New York City Approved Street Trees Suggested Tree Species Shape Visual interest Frequency of Preferred Cultivars Notes Scientific Name Common Name Planting Acer rubrum Red Maple Sparingly 'Red Sunset' ALB Host Aesculus hippocastanum Horsechestnut White May flowers Sparingly 'Baumanni' ALB Host Aesculus octandra Yellow Buckeye Yellow May Flowers Sparingly ALB Host ALB Host 'Duraheat' Betula nigra River Birch Ornamental Bark Sparingly Plant Single Stem 'Heritage' Only Celtis occidentalis Hackberry Ornamental Bark Sparingly 'Magnifica' ALB Host ALB Host Cercidiphyllum japonicum Katsura Tree Sparingly Plant Single Stem Only Corylus colurna Turkish Filbert Sparingly LARGE TREES: Mature LARGE TREES: height than greater feet 50 tall Eucommia ulmoides Hardy Rubber Tree Frequently 'Asplenifolia' Fagus sylvatica European Beech Sparingly 'Dawyckii Purple' 'Autumn Gold' Ginkgo biloba Ginkgo Yellow Fall Color Moderately 'Magyar' Very Tough Tree 'Princeton Sentry' 'Shademaster' 'Halka' Gleditsia triacanthos var inermis Honeylocust Yellow Fall Color Moderately 'Imperial' 'Skyline' 'Espresso' Gymnocladus dioicus Kentucky Coffeetree Large Tropical Leaves Frequently 'Prairie Titan' Page 1 of 7 New York City Approved Street Trees Suggested Tree Species Shape Visual interest Frequency of Preferred Cultivars Notes Scientific Name Common Name Planting 'Rotundiloba' Seedless Cultivars Liquidambar styraciflua Sweetgum Excellent Fall Color Frequently 'Worplesdon' Preffered 'Cherokee' Orange/Green June Liriodendron tulipifera Tulip Tree Moderately Flowers Metasequoia -

Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Quercus Bawanglingensis Huang, Li Et Xing, a Vulnerable Oak Tree in China
Article Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Quercus bawanglingensis Huang, Li et Xing, a Vulnerable Oak Tree in China Xue Liu 1 , Er-Mei Chang 1, Jian-Feng Liu 1,* , Yue-Ning Huang 1, Ya Wang 1, Ning Yao 1 and Ze-Ping Jiang 1,2 1 Key Laboratory of Tree Breeding and Cultivation of State Forestry Administration, Research Institute of Forestry, Chinese Academy of Forestry, Beijing 100091, China 2 Research Institute of Forest Ecology, Environment and Protection, Chinese Academy of Forestry, Beijing 100091, China * Correspondence: [email protected] Received: 5 June 2019; Accepted: 12 July 2019; Published: 15 July 2019 Abstract: Quercus bawanglingensis Huang, Li et Xing, an endemic evergreen oak of the genus Quercus (Fagaceae) in China, is currently listed in the Red List of Chinese Plants as a vulnerable (VU) plant. No chloroplast (cp) genome information is currently available for Q. bawanglingensis, which would be essential for the establishment of guidelines for its conservation and breeding. In the present study, the cp genome of Q. bawanglingensis was sequenced and assembled into double-stranded circular DNA with a length of 161,394 bp. Two inverted repeats (IRs) with a total of 51,730 bp were identified, and the rest of the sequence was separated into two single-copy regions, namely, a large single-copy (LSC) region (90,628 bp) and a small single-copy (SSC) region (19,036 bp). The genome of Q. bawanglingensis contains 134 genes (86 protein-coding genes, 40 tRNAs and eight rRNAs). More forward (29) than inverted long repeats (21) are distributed in the cp genome. -

Chapter 4 Phytogeography of Northeast Asia
Chapter 4 Phytogeography of Northeast Asia Hong QIAN 1, Pavel KRESTOV 2, Pei-Yun FU 3, Qing-Li WANG 3, Jong-Suk SONG 4 and Christine CHOURMOUZIS 5 1 Research and Collections Center, Illinois State Museum, 1011 East Ash Street, Springfield, IL 62703, USA, e-mail: [email protected]; 2 Institute of Biology and Soil Science, Russian Academy of Sciences, Vladivostok, 690022, Russia, e-mail: [email protected]; 3 Institute of Applied Ecology, Chinese Academy of Sciences, P.O. Box 417, Shenyang 110015, China; 4 Department of Biological Science, College of Natural Sciences, Andong National University, Andong 760-749, Korea, e-mail: [email protected]; 5 Department of Forest Sciences, University of British Columbia, 3041-2424 mail Mall, Vancouver, B.C., V6T 1Z4, Canada, e-mail: [email protected] Abstract: Northeast Asia as defined in this study includes the Russian Far East, Northeast China, the northern part of the Korean Peninsula, and Hokkaido Island (Japan). We determined the species richness of Northeast Asia at various spatial scales, analyzed the floristic relationships among geographic regions within Northeast Asia, and compared the flora of Northeast Asia with surrounding floras. The flora of Northeast Asia consists of 971 genera and 4953 species of native vascular plants. Based on their worldwide distributions, the 971 gen- era were grouped into fourteen phytogeographic elements. Over 900 species of vascular plants are endemic to Northeast Asia. Northeast Asia shares 39% of its species with eastern Siberia-Mongolia, 24% with Europe, 16.2% with western North America, and 12.4% with eastern North America. -

Quercus Drymeja Unger and Q. Mediterranea Unger
Review of Palaeobotany and Palynology 241 (2017) 98–128 Contents lists available at ScienceDirect Review of Palaeobotany and Palynology journal homepage: www.elsevier.com/locate/revpalbo Taxonomy and palaeoecology of two widespread western Eurasian Neogene sclerophyllous oak species: Quercus drymeja Unger and Q. mediterranea Unger Thomas Denk a,⁎, Dimitrios Velitzelos b,TuncayH.Günerc, Johannes M. Bouchal a,d, Friðgeir Grímsson d,GuidoW.Grimmd,e a Swedish Museum of Natural History, Department of Palaeobiology, Box 50007, 10405 Stockholm, Sweden b National and Kapodistrian University of Athens, Faculty of Geology and Geoenvironment, Department of Historical Geology and Paleontology, Panepistimiopolis, Athens 15784, Greece c Istanbul University, Faculty of Forestry, Department of Forest Botany, 34473 Bahceköy, Istanbul, Turkey d University of Vienna, Department of Palaeontology, 1090 Vienna, Austria e Unaffiliated, 45100 Orléans, France article info abstract Article history: Sclerophyllous oaks (genus Quercus) play important roles in Neogene ecosystems of south-western Eurasia. Received 31 May 2016 Modern analogues (‘nearest living relatives’) for these oaks have been sought among five of six infrageneric lin- Accepted 30 January 2017 eages of Quercus, distributed across the entire Northern Hemisphere. A revision of leaf fossils from lower Miocene Available online 10 February 2017 to Pliocene deposits suggests that morphotypes of the Quercus drymeja complex are very similar to a number of extant Himalayan, East Asian, and Southeast Asian species of Quercus Group Ilex and may indicate subtropical, Keywords: Quercus Group Ilex relatively humid conditions. Quercus mediterranea comprises leaf morphotypes that are encountered in modern Plant fossil Mediterranean species of Quercus Group Ilex, but also in Himalayan and East Asian members of this group indi- Modern analogue cating fully humid or summer-wet conditions. -

Wild Crop Relatives: Genomic and Breeding Resources: Forest Trees
Wild Crop Relatives: Genomic and Breeding Resources . Chittaranjan Kole Editor Wild Crop Relatives: Genomic and Breeding Resources Forest Trees Editor Prof. Chittaranjan Kole Director of Research Institute of Nutraceutical Research Clemson University 109 Jordan Hall Clemson, SC 29634 [email protected] ISBN 978-3-642-21249-9 e-ISBN 978-3-642-21250-5 DOI 10.1007/978-3-642-21250-5 Springer Heidelberg Dordrecht London New York Library of Congress Control Number: 2011922649 # Springer-Verlag Berlin Heidelberg 2011 This work is subject to copyright. All rights are reserved, whether the whole or part of the material is concerned, specifically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfilm or in any other way, and storage in data banks. Duplication of this publication or parts thereof is permitted only under the provisions of the German Copyright Law of September 9, 1965, in its current version, and permission for use must always be obtained from Springer. Violations are liable to prosecution under the German Copyright Law. The use of general descriptive names, registered names, trademarks, etc. in this publication does not imply, even in the absence of a specific statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use. Cover design: deblik, Berlin Printed on acid-free paper Springer is part of Springer Science+Business Media (www.springer.com) Dedication Dr. Norman Ernest Borlaug,1 the Father of Green Revolution, is well respected for his contribu- tions to science and society. There was or is not and never will be a single person on this Earth whose single-handed service to science could save millions of people from death due to starvation over a period of over four decades like Dr. -

Trees, Shrubs, and Perennials That Intrigue Me (Gymnosperms First
Big-picture, evolutionary view of trees and shrubs (and a few of my favorite herbaceous perennials), ver. 2007-11-04 Descriptions of the trees and shrubs taken (stolen!!!) from online sources, from my own observations in and around Greenwood Lake, NY, and from these books: • Dirr’s Hardy Trees and Shrubs, Michael A. Dirr, Timber Press, © 1997 • Trees of North America (Golden field guide), C. Frank Brockman, St. Martin’s Press, © 2001 • Smithsonian Handbooks, Trees, Allen J. Coombes, Dorling Kindersley, © 2002 • Native Trees for North American Landscapes, Guy Sternberg with Jim Wilson, Timber Press, © 2004 • Complete Trees, Shrubs, and Hedges, Jacqueline Hériteau, © 2006 They are generally listed from most ancient to most recently evolved. (I’m not sure if this is true for the rosids and asterids, starting on page 30. I just listed them in the same order as Angiosperm Phylogeny Group II.) This document started out as my personal landscaping plan and morphed into something almost unwieldy and phantasmagorical. Key to symbols and colored text: Checkboxes indicate species and/or cultivars that I want. Checkmarks indicate those that I have (or that one of my neighbors has). Text in blue indicates shrub or hedge. (Unfinished task – there is no text in blue other than this text right here.) Text in red indicates that the species or cultivar is undesirable: • Out of range climatically (either wrong zone, or won’t do well because of differences in moisture or seasons, even though it is in the “right” zone). • Will grow too tall or wide and simply won’t fit well on my property. -

The Effects of Chemical Components and Particle Size on the Mechanical
Journal of Wood Science (2018) 64:246–255 https://doi.org/10.1007/s10086-018-1695-y ORIGINAL ARTICLE The effects of chemical components and particle size on the mechanical properties of binderless boards made from oak (Quercus spp.) logs degraded by shiitake fungi (Lentinula edodes) Florence Hiu Yan Lui1 · Yoko Kurokochi1 · Hiroe Narita1 · Yukie Saito1 · Masatoshi Sato1 Received: 27 September 2017 / Accepted: 22 December 2017 / Published online: 15 February 2018 © The Japan Wood Research Society 2018 Abstract Binderless boards are composite boards that rely on self-bonding mechanisms for inter-fibre bonding. Quercus acutissima and Quercus serrata logs degraded by Lentinula edodes (shiitake fungi) were used in this study to investigate whether physical and chemical changes induced by shiitake fungi can enhance board mechanical properties. Binderless boards were manufactured with 0.8 g/cm3 target density, 220 °C pressing temperature, 5 MPa pressure, and pressing duration of 10 min. Boards made from logs degraded for ≥ 26 months were stronger than control boards and met modulus of rupture (MOR) and internal bonding (IB) requirements for fibreboards. Chemical composition and particle size distribution of the wood powder used to make the boards were determined to elucidate the drivers of board mechanical properties. The proportion of small particles (< 150 µm) showed a strong positive correlation with MOR for both species and hot water extractives showed a strong positive correlation with IB for Q. acutissima boards. Introduction of shiitake fungi pre-treatment to the production process may enhance the mechanical strength of binderless boards. Keywords Binderless board · Internal bonding · Shiitake · Degradation Introduction Research on binderless boards has coincided with the drive for agro-industrial waste recycling and the develop- Wood-based composites are produced by combining wood ment of ecomaterials. -

Earlywood Vessels in Ringsporous Trees Become Functional for Water Transport After Bud Burst and Before the Maturation of the Cu
Kitin et al.IAWA – Earlywood Journal 37vessel (2), development2016: 315–331 and function 315 EARLYWOOD VESSELS IN RING-POROUS TREES BECOME FUNCTIONAL FOR WATER TRANSPORT AFTER BUD BURST AND BEFORE THE MATURATION OF THE CURRENT-YEAR LEAVES Peter Kitin1, 2,* and Ryo Funada3 1USDA Forest Service, Forest Products Laboratory, One Gifford Pinchot Drive, Madison, WI 53726, U.S.A. 2Madison Area Technical College, 1701 Wright Street, Madison, WI 53704, U.S.A. 3Faculty of Agriculture, Tokyo University of Agriculture and Technology, Fuchu-Tokyo 183-8509, Japan *Corresponding author; e-mail: [email protected] ABSTRACT This paper reviews the development of xylem vessels in ring-porous dicots and the corresponding leaf phenology. Also included are our original observations on the time-course of vessel element growth, secondary wall deposition, and end wall perforation in the deciduous hardwood Kalopanax septemlobus. Dif- ferent patterns of xylem growth and phenology serve different strategies of the species for adaptation to seasonal climates. Trees with ring-porous xylem form wide earlywood vessels (EWV) in spring and narrow latewood vessels in sum- mer. The wide EWV become embolized or blocked with tyloses by the end of the growing season while the narrow vessels may remain functional for many years. The co-occurrence of wide and narrow vessels provides both efficiency and safety of the water transport as well as a potentially longer growing sea- son. It has for a long time been assumed that EWV in ring-porous hardwoods are formed in early spring before bud burst in order to supply sap to growing leaves and shoots. -

An Updated Infrageneric Classification of the North American Oaks
Article An Updated Infrageneric Classification of the North American Oaks (Quercus Subgenus Quercus): Review of the Contribution of Phylogenomic Data to Biogeography and Species Diversity Paul S. Manos 1,* and Andrew L. Hipp 2 1 Department of Biology, Duke University, 330 Bio Sci Bldg, Durham, NC 27708, USA 2 The Morton Arboretum, Center for Tree Science, 4100 Illinois 53, Lisle, IL 60532, USA; [email protected] * Correspondence: [email protected] Abstract: The oak flora of North America north of Mexico is both phylogenetically diverse and species-rich, including 92 species placed in five sections of subgenus Quercus, the oak clade centered on the Americas. Despite phylogenetic and taxonomic progress on the genus over the past 45 years, classification of species at the subsectional level remains unchanged since the early treatments by WL Trelease, AA Camus, and CH Muller. In recent work, we used a RAD-seq based phylogeny including 250 species sampled from throughout the Americas and Eurasia to reconstruct the timing and biogeography of the North American oak radiation. This work demonstrates that the North American oak flora comprises mostly regional species radiations with limited phylogenetic affinities to Mexican clades, and two sister group connections to Eurasia. Using this framework, we describe the regional patterns of oak diversity within North America and formally classify 62 species into nine major North American subsections within sections Lobatae (the red oaks) and Quercus (the Citation: Manos, P.S.; Hipp, A.L. An Quercus Updated Infrageneric Classification white oaks), the two largest sections of subgenus . We also distill emerging evolutionary and of the North American Oaks (Quercus biogeographic patterns based on the impact of phylogenomic data on the systematics of multiple Subgenus Quercus): Review of the species complexes and instances of hybridization. -

Nursery Raising and Plantation Techniques in Temperate (Oak) Tasar Activities
Popular Article www.vigyanvarta.com Vol-2 Issue-5 Binkadakatt and Verma (2021) Nursery Raising and Plantation Techniques in Temperate (Oak) Tasar Activities Binkadakatti, J1* and Verma, A.S2 1Scientist-C, Research Extension Centre, Central Silk Board, Palampur (Himachal Pradesh) 2Scientist-D, Regional Sericulture Research Station, Central Silk Board, Bhimtal (Uttarakhand) Corresponding Author Binkadakatti, J Email ID: [email protected] OPEN ACCESS Keywords Temperate tasar, Nursery, Quercus and Pruning How to cite this article Binkadakatti, J. and Verma, A.S. 2021. Nursery Raising and Plantation techniques in Temperate (Oak) Tasar Activities. Vigyan Varta 2(5): 1-4. ABSTRACT Oak belongs to the beech family Fagaceae, Order Fagales and genus Quercus is the only food plant of oak tasar silkworm Antheraea proylei J. Many plants commonly called Oak are not belonged to Quercus species e.g., African oak, Australian Oak, bull oak, Jerusalem oak, poison oak, river oak, she-oak, silky oak, Tasmanian oak, tulip oak, silver oak etc.Some commercial Oak food plants which were using to take Temperate (Oak) Tasar rearing activities in India are Quercus Incana, Quercus serrata, Quercus semicarpifolia, Quercus dildata, Quercus floribunda, Quercus himalayana, Quercus griffithii, Quercus leucotrichophora. Among these food plants Quercus serratais fast growing species and recommended for commercial plantation for Oak tasar activities. INTRODUCTION silkworm is a temperate species feeds on different species of the leaves of oak tree n India, tasar and oak tasar culture is Quercus species found in the states of practiced from the immemorial in tropical Arunachal Pradesh, Assam, Himachal Pradesh, I and temperate belts even prior to the Jammu and Kashmir, Manipur, Meghalaya, introduction of mulberry in India.