Phylogenetic Analysis of Two Single-Copy Nuclear Genes Revealed Origin and Complex Relationships of Hordeum Polyploids in Tritic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hordeum Murinum L. Ssp. Leporinum (Link) Arcang. USDA
NEW YORK NON-NATIVE PLANT INVASIVENESS RANKING FORM Scientific name: Hordeum murinum L. ssp. leporinum (Link) Arcang. USDA Plants Code: HOMUL Common names: leporinum barley; hare barley Native distribution: Eurasia Date assessed: July 16, 2012 Assessors: Steven D. Glenn Reviewers: LIISMA SRC Date Approved: 14 August 2012 Form version date: 29 April 2011 New York Invasiveness Rank: Not Assessable Distribution and Invasiveness Rank (Obtain from PRISM invasiveness ranking form) PRISM Status of this species in each PRISM: Current Distribution Invasiveness Rank 1 Adirondack Park Invasive Program Not Assessed Not Assessed 2 Capital/Mohawk Not Assessed Not Assessed 3 Catskill Regional Invasive Species Partnership Not Assessed Not Assessed 4 Finger Lakes Not Assessed Not Assessed 5 Long Island Invasive Species Management Area Not Present Not Assessable 6 Lower Hudson Not Assessed Not Assessed 7 Saint Lawrence/Eastern Lake Ontario Not Assessed Not Assessed 8 Western New York Not Assessed Not Assessed Invasiveness Ranking Summary Total (Total Answered*) Total (see details under appropriate sub-section) Possible 1 Ecological impact 40 (10) 3 2 Biological characteristic and dispersal ability 25 (22) 15 3 Ecological amplitude and distribution 25 (21) 8 4 Difficulty of control 10 (6) 2 Outcome score 100 (59)b 28a † Relative maximum score -- § New York Invasiveness Rank Not Assessable * For questions answered “unknown” do not include point value in “Total Answered Points Possible.” If “Total Answered Points Possible” is less than 70.00 points, then the overall invasive rank should be listed as “Unknown.” †Calculated as 100(a/b) to two decimal places. §Very High >80.00; High 70.00−80.00; Moderate 50.00−69.99; Low 40.00−49.99; Insignificant <40.00 Not Assessable: not persistent in NY, or not found outside of cultivation. -
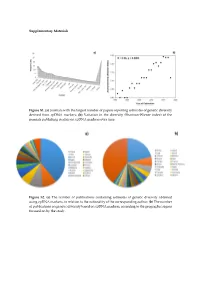
(A) Journals with the Largest Number of Papers Reporting Estimates Of
Supplementary Materials Figure S1. (a) Journals with the largest number of papers reporting estimates of genetic diversity derived from cpDNA markers; (b) Variation in the diversity (Shannon-Wiener index) of the journals publishing studies on cpDNA markers over time. Figure S2. (a) The number of publications containing estimates of genetic diversity obtained using cpDNA markers, in relation to the nationality of the corresponding author; (b) The number of publications on genetic diversity based on cpDNA markers, according to the geographic region focused on by the study. Figure S3. Classification of the angiosperm species investigated in the papers that analyzed genetic diversity using cpDNA markers: (a) Life mode; (b) Habitat specialization; (c) Geographic distribution; (d) Reproductive cycle; (e) Type of flower, and (f) Type of pollinator. Table S1. Plant species identified in the publications containing estimates of genetic diversity obtained from the use of cpDNA sequences as molecular markers. Group Family Species Algae Gigartinaceae Mazzaella laminarioides Angiospermae Typhaceae Typha laxmannii Angiospermae Typhaceae Typha orientalis Angiospermae Typhaceae Typha angustifolia Angiospermae Typhaceae Typha latifolia Angiospermae Araliaceae Eleutherococcus sessiliflowerus Angiospermae Polygonaceae Atraphaxis bracteata Angiospermae Plumbaginaceae Armeria pungens Angiospermae Aristolochiaceae Aristolochia kaempferi Angiospermae Polygonaceae Atraphaxis compacta Angiospermae Apocynaceae Lagochilus macrodontus Angiospermae Polygonaceae Atraphaxis -

An Unmöglichen Dingen Soll Man Selten Verzweifeln, an Schweren Nie
An unmöglichen Dingen soll man selten verzweifeln, an schweren nie. Johann Wolfgang von Goethe Cytogenetic mapping of BAC contigs assigned to barley chromosome 3H and comparative subchromosomal analysis within the genus Hordeum Kumulative Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften doctor rerum naturalium (Dr. rer. nat) der Naturwissenschaftlichen Fakultät III- Agrar- und Ernährungswissenschaften, Geowissenschaften und Informatik der Martin-Luther-Universität Halle-Wittenberg Vorgelegt von Lala Aliyeva-Schnorr (geb. Aliyeva) geboren am 04.04.1985 in Baku, Aserbaidschan 1. Gutachter: Prof. Dr. Klaus Pillen 2. Gutachter: Prof. Dr. Thomas Schmidt 3. Gutachter: Dr. habil. Andreas Houben Verteidigung am 21. November 2016 Acknowledgements Acknowledgements This work was performed from April 2012 till February 2016 at the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben. The financial support from IPK made the start of this project possible. Since September 2013 it was funded by German Research Foundation (DFG – Deutsche Forschungsgemeinschaft). First of all I would like to express my special thanks to Dr. habil. Andreas Houben, the head of Chromosome Structure and Function (CSF) group, for giving me the opportunity to be the part of his team, for continuous guidance, permanent encouragement as well as fruitful discussions. Also, I would like to thank Dr. Nils Stein for his help, support and discussions during my time at IPK. Special thanks to Prof. Klaus Pillen for his supportive supervision of my doctoral research and to Prof. Thomas Schmidt for the review of this thesis. My deepest thanks to all present and former members as well as visitors of the group CSF. All of them made our group an enjoyable place to work. -

Appendix C Plant and Animal Species Observed
Appendix C Plant and Animal Species Observed This list includes vascular plants, mammals, birds, reptiles, and amphibians observed in the BSA by biologists during various surveys in 2005 and 2006. This list does not include invertebrate species. Invertebrates that would be most commonly encountered on the site would include butterflies, flies, dragonflies, damselflies, beetles, earwigs, grasshoppers, crickets, termites, true bugs, mantids, lacewings, bees, wasps, ants, and spiders. PLANT SPECIES OBSERVED Scientific Name Common Name Invasive Plant Rating CLUB AND SPIKE MOSSES Selaginellaceae Spike moss family Selaginella cinerascens Mesa spikemoss TRUE FERNS Azollaceae Mosquito fern family Azolla filiculoides Pacific mosquito fern Marsileaceae Marsilea family Marsilea vestita Hairy waterclover Pilularia americana American pillwort Pteridaceae Lip fern family Pellaea andromedifolia Coffee fern Pentagramma triangularis Goldenback fern PINOPHYTA GYMNOSPERMS Cupressaceace Cypress family Juniperus californica California juniper DICOT FLOWERING PLANTS Aizoaceae Carpet weed family Carpobrotus edulis* Hottentot-fig HIGH Mesembryanthemum nodiflorum* Slender-leaved ice plant Trianthema portulacastrum Horse-purslane Amaranthaceae Amaranth family Amaranthus albus* Tumbling pigweed Amaranthus palmeri Palmer’s pigweed Amaranthus sp. Pigweed Anacardiaceae Sumac family Malosma laurina Laurel sumac Rhus ovata Sugar bush Rhus trilobata Skunkbush sumac Schinus molle* Peruvian pepper tree LIMITED Schinus terebinthifolius* Brazilian pepper tree LIMITED Mid -

Hordeum (Poaceae: Triticeae)
Breeding Science 59: 471–480 (2009) Review Progress in phylogenetic analysis and a new infrageneric classification of the barley genus Hordeum (Poaceae: Triticeae) Frank R. Blattner* Leibniz Institute of Plant Genetics and Crop Research (IPK), D-06466 Gatersleben, Germany During the last decade several phylogenetic studies of Hordeum were published using a multitude of loci from the chloroplast and nuclear genomes. In many studies taxon sampling was not representative and, thus, does not allow the inference of relationships among species. Generally, chloroplast data seem not suitable for reliable phylogenetic results, as far reaching incomplete lineage sorting result in nearly arbitrary species relationships within narrow species groups, depending on the individuals included in the analyses. Nuclear loci initially resulted at contradictory phylogenetic hypotheses. However, combining at least three nuclear loci in total evidence analyses finally provides consistent relationships among Hordeum species groups, and supports data from earlier cytological and karyological studies. Thus, recently published phylogenies agree on the monophyly of the four Hordeum genome groups (H, I, Xa, Xu), monophyly of the H/Xu and I/Xa groups, and separation between Asian and American members within the I-genome group. A new infra- generic classification of Hordeum is proposed, dividing the genus in subgenus Hordeum comprising sections Hordeum and Trichostachys, and subgenus Hordeastrum with sections Marina, Nodosa, and Stenostachys. The latter consists of two series reflecting the geographical distribution of the taxa, i.e. series Sibirica with Central Asian taxa and series Critesion comprising native taxa from the Americas. In section Nodosa all allo- polyploid taxa are grouped, which are characterized by I/Xa genome combinations. -

Structural Characteristics of Scbx Genes Controlling the Biosynthesis of Hydroxamic Acids in Rye (Secale Cereale L.)
J Appl Genetics (2015) 56:287–298 DOI 10.1007/s13353-015-0271-z PLANT GENETICS • ORIGINAL PAPER Structural characteristics of ScBx genes controlling the biosynthesis of hydroxamic acids in rye (Secale cereale L.) Beata Bakera & Bogna Makowska & Jolanta Groszyk & Michał Niziołek & Wacław Orczyk & Hanna Bolibok-Brągoszewska & Aneta Hromada-Judycka & Monika Rakoczy-Trojanowska Received: 15 July 2014 /Revised: 12 January 2015 /Accepted: 13 January 2015 /Published online: 10 February 2015 # The Author(s) 2015. This article is published with open access at Springerlink.com Abstract Benzoxazinoids (BX) are major secondary metab- analyses of the resulting contigs were used to examine the olites of gramineous plants that play an important role in dis- structure and other features of these genes, including their ease resistance and allelopathy. They also have many other promoters, introns and 3’UTRs. Comparative analysis unique properties including anti-bacterial and anti-fungal ac- showed that the ScBx genes are similar to those of other tivity, and the ability to reduce alfa–amylase activity. The bio- Poaceae species, especially to the TaBx genes. The polymor- synthesis and modification of BX are controlled by the genes phisms present both in the coding sequences and non-coding Bx1 ÷ Bx10, GT and glu, and the majority of these Bx genes regions of ScBx in relation to other Bx genes are predicted to have been mapped in maize, wheat and rye. However, the have an impact on the expression, structure and properties of genetic basis of BX biosynthesis remains largely the encoded proteins. uncharacterized apart from some data from maize and wheat. -

Hordeum Jubatum Foxtail Barley
Hordeum jubatum Foxtail Barley by Kathy Lloyd Montana Native Plant Society Photo: Drake Barton Hordeum jubatum (Foxtail Barley) label is still on the specimen sheet at the Lewis & Clark Herbarium at the Academy of Natural Sci- here are two specimens of foxtail barley ences in Philadelphia. (Hordeum jubatum) in the Lewis & Clark Foxtail barely is a native perennial bunchgrass in, THerbarium today. One of them was collected in believe it or not, the grass family (Poaceae). It is in- Montana and one at Fort Clatsop in Oregon. The digenous to the western United States but is now Montana specimen was collected on July 12, 1806 naturalized in the eastern U.S. and occurs throughout on White Bear Island in the Missouri River near pre- much of the country with the exception of the South sent-day Great Falls. Lewis and his party were on Atlantic and Gulf Coast states. It also occurs their way to explore the Marias River basin and throughout most of Canada and some areas of Mex- stopped at White Bear Island on their way. Foxtail ico. The species is considered rare in Virginia. The barley is not mentioned in Lewis’s journal entry for grass is usually less than 32 inches tall and has flat that day, but the year before, on June 25 when the blades and a hollow stem. It is distinctive because expedition was also at White Bear Island, Lewis of the long, fine, bristle-like awns on the spikelets says of this grass, “there is a species of wild rye (the “fine and soft beard” referred to by Lewis), and which is now heading it rises to the hight of 18 or 20 the fact that three spikelets join together at a com- inches, the beard is remarkably fine and soft it is a mon point. -

The Genetic Background of Benzoxazinoid Biosynthesis in Cereals
Acta Physiol Plant (2015) 37:176 DOI 10.1007/s11738-015-1927-3 REVIEW The genetic background of benzoxazinoid biosynthesis in cereals 1 1 1 B. Makowska • B. Bakera • M. Rakoczy-Trojanowska Received: 8 July 2014 / Revised: 11 May 2015 / Accepted: 22 July 2015 / Published online: 6 August 2015 Ó The Author(s) 2015. This article is published with open access at Springerlink.com Abstract Benzoxazinoids (BXs) are important com- Occurrence, characterization and biological role pounds in plant defense. Their allelopathic, nematode of BXs suppressive and antimicrobial properties are well known. BXs are found in monocot plants and in a few Benzoxazinoids (BXs) are protective and allelophatic sec- species of dicots. Over 50 years of study have led to ondary metabolites found in numerous species belonging to the characterization of the chromosomal locations and the Poaceae family, including maize, rye, wheat (Niemeyer coding sequences of almost all the genes involved in 1988a;Gru¨netal.2005;Nomuraetal.2007;Freyetal. BX biosynthesis in a number of cereal species: 2009; Chu et al. 2011; Sue et al. 2011), the wheat progen- ZmBx1–ZmBx10a7c in maize, TaBx1–TaBx5, TaGT itors Triticum urartu, Aegilops speltoides, Aegilops squar- and Taglu in wheat, ScBx17ScBx5, ScBx6-like, ScGT rosa (Niemeyer 1988b), and wild barleys: Hordeum and Scglu in rye. So far, the ortholog of the maize Bx7 roshevitzii, Hordeum flexuosum, Hordeum brachyantherum, gene has not been identified in the other investigated Hordeum lechleri (Gru¨netal.2005) and in genera—Chus- species. This review aims to summarize the available quea, Elymus, Arudo (Zu`n˜iga et al. 1983), Coix (Nagao et al. -

Large Trees, Supertrees and the Grass Phylogeny
LARGE TREES, SUPERTREES AND THE GRASS PHYLOGENY Thesis submitted to the University of Dublin, Trinity College for the Degree of Doctor of Philosophy (Ph.D.) by Nicolas Salamin Department of Botany University of Dublin, Trinity College 2002 Research conducted under the supervision of Dr. Trevor R. Hodkinson Department of Botany, University of Dublin, Trinity College Dr. Vincent Savolainen Jodrell Laboratory, Molecular Systematics Section, Royal Botanic Gardens, Kew, London DECLARATION I thereby certify that this thesis has not been submitted as an exercise for a degree at any other University. This thesis contains research based on my own work, except where otherwise stated. I grant full permission to the Library of Trinity College to lend or copy this thesis upon request. SIGNED: ACKNOWLEDGMENTS I wish to thank Trevor Hodkinson and Vincent Savolainen for all the encouragement they gave me during the last three years. They provided very useful advice on scientific papers, presentation lectures and all aspects of the supervision of this thesis. It has been a great experience to work in Ireland, and I am especially grateful to Trevor for the warm welcome and all the help he gave me, at work or outside work, since the beginning of this Ph.D. in the Botany Department. I will always remember his patience and kindness to me at this time. I am also grateful to Vincent for his help and warm welcome during the different periods of time I stayed in London, but especially for all he did for me since my B.Sc. at the University of Lausanne. I wish also to thank Prof. -

Variation of Nitrate Reductase Genes in Selected Grass Species
Variation of nitrate reductase genes in selected grass species Jizhong Zhou, Andrzej Kilian, Robert L. Warner, and Andris Kleinhofs Abstract: In order to study the variation of nitrate reductase (NR) genes among grass species, gene number, intron size and number, and the heme-hinge fragment sequence of 25 grass species were compared. Genomic DNA cut with six restriction enzymes and hybridized with the barley NAD(P)H and NADH NR gene probes revealed a single NAD(P)H NR gene copy and two or more NADH NR gene copies per haploid genome in most of the species examined. Major exceptions were Hordeum vulgare, H. vulgare ssp. spontaneum, and Avena strigosa, which appeared to have a single NADH NR gene copy. The NADH NR gene intron number and lengths were examined by polymerase chain reaction amplification. Introns I and I11 appeared to be absent in at least one of the NADH NR genes in the grass species, while intron I1 varied from 0.8 to 2.4 kilobases in length. The NADH NR gene heme-hinge regions were amplified and sequenced. The estimated average overall nucleotide substitution rate in the sequenced region was 7.8 X lo-'' substitutions/site per year. The synonymous substitution rate was 2.11 X substitutions/synonymous site per year and the nonsynonymous substitution rate was 4.10 X lo-'' substitutions/nonsynonymous site per year. Phylogenetic analyses showed that all of the wild Hordeum species examined clustered in a group separate from H. vulgare and H. vulgare ssp. spontaneum. Key words: nitrate reductase gene, gene copy number, intron, molecular phylogeny, grasses. -

Expression and Function of the Chloroplast-Encoded Gene Matk
Expression and Function of the Chloroplast-encoded Gene matK. Michelle Marie Barthet Dissertation submitted to the faculty of the Virginia Polytechnic Institute and State University in partial fulfillment of the requirements for the degree of Doctor of Philosophy In Biological Sciences K. W. Hilu, Chair E. Beers G. Gillaspy J. Sible R. A. Walker February 9, 2006 Blacksburg, Virginia Keywords: MatK, chloroplast, maturase, fast-evolving, Orchidaceae Copyright 2006, Michelle Marie Barthet Expression and Function of the chloroplast-encoded gene matK. Michelle Marie Barthet ABSTRACT The chloroplast matK gene has been identified as a rapidly evolving gene at nucleotide and corresponding amino acid levels. The high number of nucleotide substitutions and length mutations in matK has provided a strong phylogenetic signal for resolving plant phylogenies at various taxonomic levels. However, these same features have raised questions as to whether matK produces a functional protein product. matK is the only proposed chloroplast-encoded group II intron maturase. There are 15 genes in the chloroplast that would require a maturase for RNA splicing. Six of these genes have introns that are not excised by a nuclear imported maturase, leaving MatK as the only candidate for processing introns in these genes. Very little research has been conducted concerning the expression and function of this important gene and its protein product. It has become crucial to understand matK expression in light of its significance in RNA processing and plant systematics. In this study, we examined the expression, function and evolution of MatK using a combination of molecular and genetic methods. Our findings indicate that matK RNA and protein is expressed in a variety of plant species, and expression of MatK protein is regulated by development. -

Cereal Rust Bulletins St
CEREAL RUST Report No. 9 BULLETIN July 22, 2003 Issued by: For the latest cereal rust news from the field, subscribe to the cereal-rust-survey mail list. To subscribe, send an email Cereal Disease Laboratory message with the word subscribe in the message body U.S. Department of Agriculture (not subject line) to: [email protected] Agricultural Research Service 1551 Lindig St, University of Minnesota Reports from this mail list as well as all Cereal Rust Bulletins St. Paul, MN 55108-6052 are maintained on the CDL web page (612) 625-6299 FAX (651) 649-5054 (http://www.cdl.umn.edu/). [email protected] • • Wheat leaf rust in the northern plains is increasing in severity on spring wheat cultivars. • • Viable wheat stripe rust is still present in the northern Great Plains. • • Oat stem rust and oat crown rust is common in upper Midwest fields. • • Stem rust is common on many wild grasses in southeastern Minnesota. The small grain harvest has commenced from southwestern New York to South Dakota. Winter wheat is in good condition and at normal maturity throughout much of the U.S. In the northern small grain area, most of the spring-sown grains are in good condition and slightly behind normal crop development. Wheat stem rust. In mid-July, traces of wheat stem rust were observed on the susceptible spring wheat cultivar Baart in south central Minnesota plots. The incidence of wheat stem rust infections have been lighter than normal in the northern Great Plains since little wheat stem rust developed in the southern and central U.S.