Kinetid Structure of Choanoflagellates and Choanocytes of Sponges Does
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Architecture of Cell Differentiation in Choanoflagellates And
bioRxiv preprint doi: https://doi.org/10.1101/452185; this version posted October 29, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 The architecture of cell differentiation in choanoflagellates 2 and sponge choanocytes 3 Davis Laundon1,2,6, Ben Larson3, Kent McDonald3, Nicole King3,4 and Pawel 4 Burkhardt1,5* 5 1 Marine Biological Association of the United Kingdom, The Laboratory, Citadel Hill, 6 Plymouth, PL1 2PB, United Kingdom 7 2 Plymouth University, Drake Circus, Plymouth, PL4 8AA, United Kingdom 8 3 Department of Molecular and Cell Biology, University of California, Berkeley, USA 9 4 Howard Hughes Medical Institute 10 5 Sars International Centre for Molecular Marine Biology, University of Bergen, 11 Thormohlensgate 55, 5020 Bergen, Norway 12 6 Current Affiliation: University of East Anglia, Norwich, NR4 7TJ, United Kingdom 13 14 *Correspondence [email protected] 15 16 17 18 19 20 21 22 23 24 25 bioRxiv preprint doi: https://doi.org/10.1101/452185; this version posted October 29, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 26 SUMMARY 27 Collar cells are ancient animal cell types which are conserved across the animal 28 kingdom [1] and their closest relatives, the choanoflagellates [2]. -

1 Lessons from Simple Marine Models on the Bacterial Regulation
bioRxiv preprint doi: https://doi.org/10.1101/211797; this version posted October 31, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Lessons from simple marine models on the bacterial regulation of eukaryotic development Arielle Woznicaa and Nicole Kinga,1 a Howard Hughes Medical Institute and Department of Molecular and Cell Biology, University of California, Berkeley, California 94720, United States 1 Corresponding author, [email protected]. 1 bioRxiv preprint doi: https://doi.org/10.1101/211797; this version posted October 31, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Highlights 2 - Cues from environmental bacteria influence the development of many marine 3 eukaryotes 4 5 - The molecular cues produced by environmental bacteria are structurally diverse 6 7 - Eukaryotes can respond to many different environmental bacteria 8 9 - Some environmental bacteria act as “information hubs” for diverse eukaryotes 10 11 - Experimentally tractable systems, like the choanoflagellate S. rosetta, promise to 12 reveal molecular mechanisms underlying these interactions 13 14 Abstract 15 Molecular cues from environmental bacteria influence important developmental 16 decisions in diverse marine eukaryotes. Yet, relatively little is understood about the 17 mechanisms underlying these interactions, in part because marine ecosystems are 18 dynamic and complex. -

Download This Publication (PDF File)
PUBLIC LIBRARY of SCIENCE | plosgenetics.org | ISSN 1553-7390 | Volume 2 | Issue 12 | DECEMBER 2006 GENETICS PUBLIC LIBRARY of SCIENCE www.plosgenetics.org Volume 2 | Issue 12 | DECEMBER 2006 Interview Review Knight in Common Armor: 1949 Unraveling the Genetics 1956 An Interview with Sir John Sulston e225 of Human Obesity e188 Jane Gitschier David M. Mutch, Karine Clément Research Articles Natural Variants of AtHKT1 1964 The Complete Genome 2039 Enhance Na+ Accumulation e210 Sequence and Comparative e206 in Two Wild Populations of Genome Analysis of the High Arabidopsis Pathogenicity Yersinia Ana Rus, Ivan Baxter, enterocolitica Strain 8081 Balasubramaniam Muthukumar, Nicholas R. Thomson, Sarah Jeff Gustin, Brett Lahner, Elena Howard, Brendan W. Wren, Yakubova, David E. Salt Matthew T. G. Holden, Lisa Crossman, Gregory L. Challis, About the Cover Drosophila SPF45: A Bifunctional 1974 Carol Churcher, Karen The jigsaw image of representatives Protein with Roles in Both e178 Mungall, Karen Brooks, Tracey of various lines of eukaryote evolution Splicing and DNA Repair Chillingworth, Theresa Feltwell, refl ects the current lack of consensus as Ahmad Sami Chaouki, Helen K. Zahra Abdellah, Heidi Hauser, to how the major branches of eukaryotes Salz Kay Jagels, Mark Maddison, fi t together. The illustrations from upper Sharon Moule, Mandy Sanders, left to bottom right are as follows: a single Mammalian Small Nucleolar 1984 Sally Whitehead, Michael A. scale from the surface of Umbellosphaera; RNAs Are Mobile Genetic e205 Quail, Gordon Dougan, Julian Amoeba, the large amoeboid organism Elements Parkhill, Michael B. Prentice used as an introduction to protists for Michel J. Weber many school children; Euglena, the iconic Low Levels of Genetic 2052 fl agellate that is often used to challenge Soft Sweeps III: The Signature 1998 Divergence across e215 ideas of plants (Euglena has chloroplasts) of Positive Selection from e186 Geographically and and animals (Euglena moves); Stentor, Recurrent Mutation Linguistically Diverse one of the larger ciliates; Cacatua, the Pleuni S. -

The Closest Unicellular Relatives of Animals
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Current Biology, Vol. 12, 1773–1778, October 15, 2002, 2002 Elsevier Science Ltd. All rights reserved. PII S0960-9822(02)01187-9 The Closest Unicellular Relatives of Animals B.F. Lang,1,2 C. O’Kelly,1,3 T. Nerad,4 M.W. Gray,1,5 Results and Discussion and G. Burger1,2,6 1The Canadian Institute for Advanced Research The evolution of the Metazoa from single-celled protists Program in Evolutionary Biology is an issue that has intrigued biologists for more than a 2 De´ partement de Biochimie century. Early morphological and more recent ultra- Universite´ de Montre´ al structural and molecular studies have converged in sup- Succursale Centre-Ville porting the now widely accepted view that animals are Montre´ al, Que´ bec H3C 3J7 related to Fungi, choanoflagellates, and ichthyosporean Canada protists. However, controversy persists as to the spe- 3 Bigelow Laboratory for Ocean Sciences cific evolutionary relationships among these major P.O. Box 475 groups. This uncertainty is reflected in the plethora of 180 McKown Point Road published molecular phylogenies that propose virtually West Boothbay Harbor, Maine 04575 all of the possible alternative tree topologies involving 4 American Type Culture Collection Choanoflagellata, Fungi, Ichthyosporea, and Metazoa. 10801 University Boulevard For example, a monophyletic MetazoaϩChoanoflagel- Manassas, Virginia 20110 lata group has been suggested on the basis of small 5 Department of Biochemistry subunit (SSU) rDNA sequences [1, 6, 7]. Other studies and Molecular Biology using the same sequences have allied Choanoflagellata Dalhousie University with the Fungi [8], placed Choanoflagellata prior to the Halifax, Nova Scotia B3H 4H7 divergence of animals and Fungi [9], or even placed Canada them prior to the divergence of green algae and land plants [10]. -

Fungal Evolution: Major Ecological Adaptations and Evolutionary Transitions
Biol. Rev. (2019), pp. 000–000. 1 doi: 10.1111/brv.12510 Fungal evolution: major ecological adaptations and evolutionary transitions Miguel A. Naranjo-Ortiz1 and Toni Gabaldon´ 1,2,3∗ 1Department of Genomics and Bioinformatics, Centre for Genomic Regulation (CRG), The Barcelona Institute of Science and Technology, Dr. Aiguader 88, Barcelona 08003, Spain 2 Department of Experimental and Health Sciences, Universitat Pompeu Fabra (UPF), 08003 Barcelona, Spain 3ICREA, Pg. Lluís Companys 23, 08010 Barcelona, Spain ABSTRACT Fungi are a highly diverse group of heterotrophic eukaryotes characterized by the absence of phagotrophy and the presence of a chitinous cell wall. While unicellular fungi are far from rare, part of the evolutionary success of the group resides in their ability to grow indefinitely as a cylindrical multinucleated cell (hypha). Armed with these morphological traits and with an extremely high metabolical diversity, fungi have conquered numerous ecological niches and have shaped a whole world of interactions with other living organisms. Herein we survey the main evolutionary and ecological processes that have guided fungal diversity. We will first review the ecology and evolution of the zoosporic lineages and the process of terrestrialization, as one of the major evolutionary transitions in this kingdom. Several plausible scenarios have been proposed for fungal terrestralization and we here propose a new scenario, which considers icy environments as a transitory niche between water and emerged land. We then focus on exploring the main ecological relationships of Fungi with other organisms (other fungi, protozoans, animals and plants), as well as the origin of adaptations to certain specialized ecological niches within the group (lichens, black fungi and yeasts). -

Bacterial Cues Regulate Multicellular Development and Mating in the Choanoflagellate, S
Bacterial cues regulate multicellular development and mating in the choanoflagellate, S. rosetta By Arielle Woznica A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Molecular and Cell Biology in the Graduate Division of the University of California, Berkeley Committee in charge: Professor Nicole King, Chair Professor Russell Vance Professor Diana Bautista Professor Brian Staskawicz Spring 2017 Abstract Bacterial cues regulate multicellular development and mating in the choanoflagellate, S. rosetta By Arielle Woznica Doctor of Philosophy in Molecular and Cell Biology University of California, Berkeley Professor Nicole King, Chair Animals first diverged from their unicellular ancestors in oceans dominated by bacteria, and have lived in close association with bacteria ever since. Interactions with bacteria critically shape diverse aspects of animal biology today, including developmental processes that were long thought to be autonomous. Yet, the multicellularity of animals and the often-complex communities of bacteria with which they are associated make it challenging to characterize the mechanisms underlying many bacterial-animal interactions. Thus, developing experimentally tractable host-microbe model systems will be essential for revealing the molecules and mechanisms by which bacteria influence animal development. The choanoflagellate Salpingoeca rosetta, one of the closest living relatives of animals, has emerged as an attractive model for studying host-microbe interactions. Like all choanoflagellates, S. rosetta feeds on bacteria; however, we have found that interactions between S. rosetta and bacteria extend beyond those of predator and prey. In fact, two key transitions in the life history of S. rosetta, multicellular “rosette” development and sexual reproduction, are regulated by environmental bacteria. -

Multigene Eukaryote Phylogeny Reveals the Likely Protozoan Ancestors of Opis- Thokonts (Animals, Fungi, Choanozoans) and Amoebozoa
Accepted Manuscript Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opis- thokonts (animals, fungi, choanozoans) and Amoebozoa Thomas Cavalier-Smith, Ema E. Chao, Elizabeth A. Snell, Cédric Berney, Anna Maria Fiore-Donno, Rhodri Lewis PII: S1055-7903(14)00279-6 DOI: http://dx.doi.org/10.1016/j.ympev.2014.08.012 Reference: YMPEV 4996 To appear in: Molecular Phylogenetics and Evolution Received Date: 24 January 2014 Revised Date: 2 August 2014 Accepted Date: 11 August 2014 Please cite this article as: Cavalier-Smith, T., Chao, E.E., Snell, E.A., Berney, C., Fiore-Donno, A.M., Lewis, R., Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa, Molecular Phylogenetics and Evolution (2014), doi: http://dx.doi.org/10.1016/ j.ympev.2014.08.012 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. 1 1 Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts 2 (animals, fungi, choanozoans) and Amoebozoa 3 4 Thomas Cavalier-Smith1, Ema E. Chao1, Elizabeth A. Snell1, Cédric Berney1,2, Anna Maria 5 Fiore-Donno1,3, and Rhodri Lewis1 6 7 1Department of Zoology, University of Oxford, South Parks Road, Oxford OX1 3PS, UK. -

Towards a Management Hierarchy (Classification) for the Catalogue of Life
TOWARDS A MANAGEMENT HIERARCHY (CLASSIFICATION) FOR THE CATALOGUE OF LIFE Draft Discussion Document Rationale The Catalogue of Life partnership, comprising Species 2000 and ITIS (Integrated Taxonomic Information System), has the goal of achieving a comprehensive catalogue of all known species on Earth by the year 2011. The actual number of described species (after correction for synonyms) is not presently known but estimates suggest about 1.8 million species. The collaborative teams behind the Catalogue of Life need an agreed standard classification for these 1.8 million species, i.e. a working hierarchy for management purposes. This discussion document is intended to highlight some of the issues that need clarifying in order to achieve this goal beyond what we presently have. Concerning Classification Life’s diversity is classified into a hierarchy of categories. The best-known of these is the Kingdom. When Carl Linnaeus introduced his new “system of nature” in the 1750s ― Systema Naturae per Regna tria naturae, secundum Classes, Ordines, Genera, Species …) ― he recognised three kingdoms, viz Plantae, Animalia, and a third kingdom for minerals that has long since been abandoned. As is evident from the title of his work, he introduced lower-level taxonomic categories, each successively nested in the other, named Class, Order, Genus, and Species. The most useful and innovative aspect of his system (which gave rise to the scientific discipline of Systematics) was the use of the binominal, comprising genus and species, that uniquely identified each species of organism. Linnaeus’s system has proven to be robust for some 250 years. The starting point for botanical names is his Species Plantarum, published in 1753, and that for zoological names is the tenth edition of the Systema Naturae published in 1758. -

Notophthalmus Viridescens) by a New Species of Amphibiocystidium, a Genus of Fungus-Like Mesomycetozoan Parasites Not Previously Reported in North America
203 Widespread infection of the Eastern red-spotted newt (Notophthalmus viridescens) by a new species of Amphibiocystidium, a genus of fungus-like mesomycetozoan parasites not previously reported in North America T. R. RAFFEL1,2*, T. BOMMARITO 3, D. S. BARRY4, S. M. WITIAK5 and L. A. SHACKELTON1 1 Center for Infectious Disease Dynamics, Biology Department, Penn State University, University Park, PA 16802, USA 2 Department of Biology, University of South Florida, Tampa, FL 33620, USA 3 Cooperative Wildlife Research Lab, Department of Zoology, Southern Illinois University, Carbondale, IL 62901, USA 4 Department of Biological Sciences, Marshall University, Huntington, WV 25755, USA 5 Department of Plant Pathology, Penn State University, University Park, PA 16802, USA (Received 21 March 2007; revised 17 August 2007; accepted 20 August 2007; first published online 12 October 2007) SUMMARY Given the worldwide decline of amphibian populations due to emerging infectious diseases, it is imperative that we identify and address the causative agents. Many of the pathogens recently implicated in amphibian mortality and morbidity have been fungal or members of a poorly understood group of fungus-like protists, the mesomycetozoans. One mesomycetozoan, Amphibiocystidium ranae, is known to infect several European amphibian species and was associated with a recent decline of frogs in Italy. Here we present the first report of an Amphibiocystidium sp. in a North American amphibian, the Eastern red-spotted newt (Notophthalmus viridescens), and characterize it as the new species A. viridescens in the order Dermocystida based on morphological, geographical and phylogenetic evidence. We also describe the widespread and seasonal distribution of this parasite in red-spotted newt populations and provide evidence of mortality due to infection. -
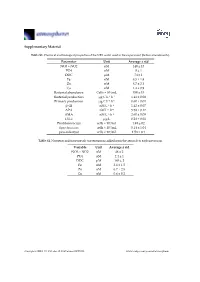
Supplementary Material Parameter Unit Average ± Std NO3 + NO2 Nm
Supplementary Material Table S1. Chemical and biological properties of the NRS water used in the experiment (before amendments). Parameter Unit Average ± std NO3 + NO2 nM 140 ± 13 PO4 nM 8 ± 1 DOC μM 74 ± 1 Fe nM 8.5 ± 1.8 Zn nM 8.7 ± 2.1 Cu nM 1.4 ± 0.9 Bacterial abundance Cells × 104/mL 350 ± 15 Bacterial production μg C L−1 h−1 1.41 ± 0.08 Primary production μg C L−1 h−1 0.60 ± 0.01 β-Gl nM L−1 h−1 1.42 ± 0.07 APA nM L−1 h−1 5.58 ± 0.17 AMA nM L−1·h−1 2.60 ± 0.09 Chl-a μg/L 0.28 ± 0.01 Prochlorococcus cells × 104/mL 1.49 ± 02 Synechococcus cells × 104/mL 5.14 ± 1.04 pico-eukaryot cells × 103/mL 1.58 × 0.1 Table S2. Nutrients and trace metals concentrations added from the aerosols to each mesocosm. Variable Unit Average ± std NO3 + NO2 nM 48 ± 2 PO4 nM 2.4 ± 1 DOC μM 165 ± 2 Fe nM 2.6 ± 1.5 Zn nM 6.7 ± 2.5 Cu nM 0.6 ± 0.2 Atmosphere 2019, 10, 358; doi:10.3390/atmos10070358 www.mdpi.com/journal/atmosphere Atmosphere 2019, 10, 358 2 of 6 Table S3. ANOVA test results between control, ‘UV-treated’ and ‘live-dust’ treatments at 20 h or 44 h, with significantly different values shown in bold. ANOVA df Sum Sq Mean Sq F Value p-value Chl-a 20 H 2, 6 0.03, 0.02 0.02, 0 4.52 0.0634 44 H 2, 6 0.02, 0 0.01, 0 23.13 0.002 Synechococcus Abundance 20 H 2, 7 8.23 × 107, 4.11 × 107 4.11 × 107, 4.51 × 107 0.91 0.4509 44 H 2, 7 5.31 × 108, 6.97 × 107 2.65 × 108, 1.16 × 107 22.84 0.0016 Prochlorococcus Abundance 20 H 2, 8 4.22 × 107, 2.11 × 107 2.11 × 107, 2.71 × 106 7.77 0.0216 44 H 2, 8 9.02 × 107, 1.47 × 107 4.51 × 107, 2.45 × 106 18.38 0.0028 Pico-eukaryote -

Protistology Molecular Phylogeny of Aphelidium Arduennense Sp. Nov
Protistology 13 (4), 192–198 (2019) Protistology Molecular phylogeny of Aphelidium arduennense sp. nov. – new representative of Aphelida (Opis- thosporidia) Victoria S. Tcvetkova1, Natalia A. Zorina1, Maria A. Mamkaeva1 and Sergey A. Karpov1,2 1 St. Petersburg State University, St. Petersburg 199034, Russia 2 Zoological Institute, Russian Academy of Sciences, St. Petersburg 199034, Russia | Submitted November 14, 2019 | Accepted December 2, 2019 | Summary Aphelids (Aphelida) are poorly known parasitoids of algae that have raised considerable interest because of their phylogenetic position as phagotrophic protists sister to Fungi. Together with Rozellida and Microsporidia they have been classified in the Opisthosporidia but seem to be more closely related to the Fungi rather than to the Cryptomycota and Microsporidia, the other members of the Opisthosporidia. Molecular environmental studies have revealed high genetic diversity within the aphelids, but only four genera have been described: Aphelidium, Amoeboaphelidium, Paraphelidium and Pseudaphelidium. Here, we describe the life cycle of a new species of Aphelidium, Aph. arduennense. Molecular phylogenetic analysis of its 18S rRNA indicates that Aph. arduennense is sister to Aph. tribonematis, and together with Aph. melosirae they form a monophyletic cluster. Within the aphelids, this cluster is distantly related to Paraphelidium and Amoeboaphelidium. Key words: aphelids, Holomycota, Opisthosporidia, Rozellosporidia, taxonomy Introduction Rozellosporidia (Cryptomycota) formed the super- phylum Opisthosporidia, the deepest branch Aphelids are a divergent group of intracellular of the Holomycota lineage, separated from the parasitoids of green, yellow-green and diatom algae Fungi (Karpov et al., 2014a; Letcher et al., 2015; (Gromov, 2000; Karpov et al., 2014a). The four 2017; Torruella et al., 2015). Several biological known genera have different ecological preferences: peculiarities of the aphelids do not conform the Aphelidium, Amoeboaphelidium and Paraphelidium classical definition of the Fungi. -

General Introduction and Objectives 3
General introduction and objectives 3 General introduction: General body organization: The phylum Porifera is commonly referred to as sponges. The phylum, that comprises more than 6,000 species, is divided into three classes: Calcarea, Hexactinellida and Demospongiae. The latter class contains more than 85% of the living species. They are predominantly marine, with the notable exception of the family Spongillidae, an extant group of freshwater demosponges whose fossil record begins in the Cretaceous. Sponges are ubiquitous benthic creatures, found at all latitudes beneath the world's oceans, and from the intertidal to the deep-sea. Sponges are considered as the most basal phylum of metazoans, since most of their features appear to be primitive, and it is widely accepted that multicellular animals consist of a monophyletic group (Zrzavy et al. 1998). Poriferans appear to be diploblastic (Leys 2004; Maldonado 2004), although the two cellular sheets are difficult to homologise with those of the rest of metazoans. They are sessile animals, though it has been shown that some are able to move slowly (up to 4 mm per day) within aquaria (e.g., Bond and Harris 1988; Maldonado and Uriz 1999). They lack organs, possessing cells that develop great number of functions. The sponge body is lined by a pseudoepithelial layer of flat cells (exopinacocytes). Anatomically and physiologically, tissues of most sponges (but carnivorous sponges) are organized around an aquiferous system of excurrent and incurrent canals (Rupert and Barnes 1995). These canals are lined by a pseudoepithelial layer of flat cells (endopinacocytes).Water flows into the sponge body through multiple apertures (ostia) General introduction and objectives 4 to the incurrent canals which end in the choanocyte chambers (Fig.