Transcriptome Profiling Analysis of Sex-Based
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Targeting and Enhancing Anticancer Efficacy of Oncolytic HSV-1 to Midkine Expressing Tumors
University of Cincinnati Date: 12/20/2010 I, Arturo R Maldonado , hereby submit this original work as part of the requirements for the degree of Doctor of Philosophy in Developmental Biology. It is entitled: Molecular Targeting and Enhancing Anticancer Efficacy of Oncolytic HSV-1 to Midkine Expressing Tumors Student's name: Arturo R Maldonado This work and its defense approved by: Committee chair: Jeffrey Whitsett Committee member: Timothy Crombleholme, MD Committee member: Dan Wiginton, PhD Committee member: Rhonda Cardin, PhD Committee member: Tim Cripe 1297 Last Printed:1/11/2011 Document Of Defense Form Molecular Targeting and Enhancing Anticancer Efficacy of Oncolytic HSV-1 to Midkine Expressing Tumors A dissertation submitted to the Graduate School of the University of Cincinnati College of Medicine in partial fulfillment of the requirements for the degree of DOCTORATE OF PHILOSOPHY (PH.D.) in the Division of Molecular & Developmental Biology 2010 By Arturo Rafael Maldonado B.A., University of Miami, Coral Gables, Florida June 1993 M.D., New Jersey Medical School, Newark, New Jersey June 1999 Committee Chair: Jeffrey A. Whitsett, M.D. Advisor: Timothy M. Crombleholme, M.D. Timothy P. Cripe, M.D. Ph.D. Dan Wiginton, Ph.D. Rhonda D. Cardin, Ph.D. ABSTRACT Since 1999, cancer has surpassed heart disease as the number one cause of death in the US for people under the age of 85. Malignant Peripheral Nerve Sheath Tumor (MPNST), a common malignancy in patients with Neurofibromatosis, and colorectal cancer are midkine- producing tumors with high mortality rates. In vitro and preclinical xenograft models of MPNST were utilized in this dissertation to study the role of midkine (MDK), a tumor-specific gene over- expressed in these tumors and to test the efficacy of a MDK-transcriptionally targeted oncolytic HSV-1 (oHSV). -
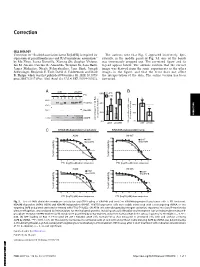
Correction for Zhou Et Al., Scaffold Association Factor B (SAFB) Is Required for Expression of Prenyltransferases and RAS Membra
Correction CELL BIOLOGY Correction for “Scaffold association factor B (SAFB) is required for The authors note that Fig. 5 appeared incorrectly. Spe- expression of prenyltransferases and RAS membrane association,” cifically, in the middle panel of Fig. 5A, one of the bands by Mo Zhou, Leena Kuruvilla, Xiarong Shi, Stephen Viviano, was erroneously cropped out. The corrected figure and its Ian M. Ahearn, Caroline R. Amendola, Wenjuan Su, Sana Badri, legend appear below. The authors confirm that the correct James Mahaffey, Nicole Fehrenbacher, Jane Skok, Joseph image was derived from the same experiments as the other Schlessinger, Benjamin E. Turk, David A. Calderwood, and Mark images in the figure, and that the error does not affect R. Philips, which was first published November 30, 2020; 10.1073/ the interpretation of the data. The online version has been pnas.2005712117 (Proc. Natl. Acad. Sci. U.S.A. 117,31914–31922). corrected. A shNT shSAFB * B 60% - + - + FTI S P S P S P S P Total Ras 40% 100% ** RhoGDI 20% 50% CORRECTION CIMPR in cytosolic fraction FTI treated vs untreated vs untreated FTI treated percentage of total Ras percentage 0% 0% shNT shSAFB GTP-loaded Ras in A549 stables, shNT shSAFB C KRAS4B-dependent lung tumor KRAS4B-independent lung A549 H1437 %cell viability %cell viability FTI (log10, µM) dose response FTI (log10, µM) dose response H358 H1975 %cell viability %cell viability FTI (log10, µM) dose response FTI (log10, µM) dose response Fig. 5. Loss of SAFB diminishes membrane association and GTP loading of KRAS4B and sensitizes KRAS4B-dependent lung tumor cells to FTI treatment. -

Mouse Skull Mean Shape and Shape Robustness Rely on Different
Mouse Skull Mean Shape and Shape Robustness Rely on Different Genetic Architectures and Different Loci Ceferino Varón-González, Luisa Pallares, Vincent Debat, Nicolas Navarro To cite this version: Ceferino Varón-González, Luisa Pallares, Vincent Debat, Nicolas Navarro. Mouse Skull Mean Shape and Shape Robustness Rely on Different Genetic Architectures and Different Loci. Frontiers inGe- netics, Frontiers, 2019, 10, pp.64. 10.3389/fgene.2019.00064. hal-02049215 HAL Id: hal-02049215 https://hal.sorbonne-universite.fr/hal-02049215 Submitted on 26 Feb 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. fgene-10-00064 February 9, 2019 Time: 17:37 # 1 ORIGINAL RESEARCH published: 12 February 2019 doi: 10.3389/fgene.2019.00064 Mouse Skull Mean Shape and Shape Robustness Rely on Different Genetic Architectures and Different Loci Ceferino Varón-González1,2, Luisa F. Pallares3, Vincent Debat1† and Nicolas Navarro2,4*† 1 Institut de Systématique, Évolution, Biodiversité, ISYEB – UMR 7205 – CNRS, MNHN, UPMC, EPHE, UA, Muséum National d’Histoire Naturelle, Sorbonne Universités, Paris, France, 2 Biogéosciences, UMR 6282 CNRS, Université Bourgogne Franche-Comté, Dijon, France, 3 Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ, United States, 4 EPHE, PSL University, Dijon, France The genetic architecture of skull shape has been extensively studied in mice and the results suggest a highly polygenic and additive basis. -
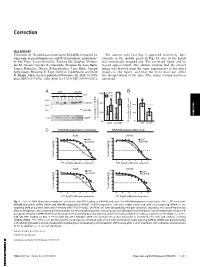
Correction for Zhou Et Al., Scaffold Association Factor B (SAFB) Is Required for Expression of Prenyltransferases and RAS Membra
Correction CELL BIOLOGY Correction for “Scaffold association factor B (SAFB) is required for The authors note that Fig. 5 appeared incorrectly. Spe- expression of prenyltransferases and RAS membrane association,” cifically, in the middle panel of Fig. 5A, one of the bands by Mo Zhou, Leena Kuruvilla, Xiarong Shi, Stephen Viviano, was erroneously cropped out. The corrected figure and its Ian M. Ahearn, Caroline R. Amendola, Wenjuan Su, Sana Badri, legend appear below. The authors confirm that the correct James Mahaffey, Nicole Fehrenbacher, Jane Skok, Joseph image was derived from the same experiments as the other Schlessinger, Benjamin E. Turk, David A. Calderwood, and Mark images in the figure, and that the error does not affect R. Philips, which was first published November 30, 2020; 10.1073/ the interpretation of the data. The online version has been pnas.2005712117 (Proc. Natl. Acad. Sci. U.S.A. 117,31914–31922). corrected. A shNT shSAFB * B 60% - + - + FTI S P S P S P S P Total Ras 40% 100% ** RhoGDI 20% 50% CORRECTION CIMPR in cytosolic fraction FTI treated vs untreated vs untreated FTI treated percentage of total Ras percentage 0% 0% shNT shSAFB GTP-loaded Ras in A549 stables, shNT shSAFB C KRAS4B-dependent lung tumor KRAS4B-independent lung A549 H1437 %cell viability %cell viability FTI (log10, µM) dose response FTI (log10, µM) dose response H358 H1975 %cell viability %cell viability FTI (log10, µM) dose response FTI (log10, µM) dose response Fig. 5. Loss of SAFB diminishes membrane association and GTP loading of KRAS4B and sensitizes KRAS4B-dependent lung tumor cells to FTI treatment. -
Evaluation of Variability in High Resolution Protein Structures by Global Distance Scoring
bioRxiv preprint doi: https://doi.org/10.1101/202028; this version posted December 16, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Evaluation of variability in high resolution protein structures by global distance scoring Risa Anzai, Yoshiki Asami, Waka Inoue, Hina Ueno, Koya Yamada, Tetsuji Okada Department of Life Science, Gakushuin University, 1-5-1 Mejiro, Toshima-ku, Tokyo 171-8588, Japan. Corresponding author: Tetsuji Okada, Department of Life Science, Gakushuin University, 1-5-1 Mejiro, Toshima-ku, Tokyo 171-8588, Japan. +81-3-3986-0221, [email protected] Abstract Systematic analysis of statistical and dynamical properties of proteins is critical to understanding cellular events. Extraction of biologically relevant information from a set of high-resolution structures is important because it can provide mechanistic details behind the functional properties of protein families, enabling rational comparison between families. Most of the current structure comparisons are pairwise-based, which hampers the global analysis of increasing contents in the Protein Data Bank. Additionally, pairing of protein structures introduces uncertainty with respect to reproducibility because it frequently accompanies other settings for superimposition. This study introduces intramolecular distance scoring, for the analysis of human proteins, for each of which at least several high-resolution are available. We show that the results are comprehensively used to overview advances at the atomic level exploration of each protein and protein family. -

A Gene for Universal Congenital Alopecia Maps to Chromosome 8P21-22 Markus M
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Am. J. Hum. Genet. 62:386–390, 1998 A Gene for Universal Congenital Alopecia Maps to Chromosome 8p21-22 Markus M. No¨then,1 Sven Cichon,1 Ina R. Vogt,1 Susanne Hemmer,1 Roland Kruse,1 Michael Knapp,2 Tobias Ho¨ller,2 Muhammad Faiyaz ul Haque,4,5 Sayedul Haque,4 Peter Propping,1 Mahmud Ahmad,4 and Marcella Rietschel1,3 1Institute of Human Genetics, 2Institute for Medical Statistics, and 3Department of Psychiatry, University of Bonn, Bonn; 4Department of Biological Sciences, Quaid-i-Azam University, Islamabad, Pakistan; and 5Medical Genetics, Ahmanson Department of Pediatrics, Cedars-Sinai Research Institute, Los Angeles Summary alopecia may occur either in isolation or with associated defects. On the basis of such associations, several dif- Complete or partial congenital absence of hair (congen- ferent syndromes featuring congenital alopecia can be ital alopecia) may occur either in isolation or with as- distinguished (Birnbaum and Baden 1987; Feinstein et sociated defects. The majority of families with isolated al. 1987; Vogt et al. 1988). The isolated form of con- congenital alopecia has been reported to follow an au- genital alopecia has been reported in sporadic and fa- tosomal-recessive mode of inheritance (MIM 203655). milial cases. In familial cases, inheritance is usu- As yet, no gene has been linked to isolated congenital ally autosomal recessive (MIM 203655 [http:// alopecia, nor has linkage been established to a specific www3.ncbi.nlm.nih.gov:80/htbin-post/Omim/dispmim? region of the genome. -

Analysis of Protein-Protein Interaction Byin Vivo Quantitative Proteomics In
Analysis of protein-protein interaction by in vivo quantitative proteomics in Caenorhabditis elegans D i s s e r t a t i o n zur Erlangung des akademischen Grades d o c t o r r e r u m n a t u r a l i u m (Dr. rer. nat.) im Fach Biologie eingereicht an der Lebenswissenschaftlichen Fakultät der Humboldt-Universität zu Berlin von M.Sc. Jiaxuan Chen Präsident der Humboldt-Universität zu Berlin Prof. Dr. Jan-Hendrik Olbertz Dekan der Lebenswissenschaftlichen Fakultät Prof. Dr. Richard Lucius Gutachter/innen: 1. Prof. Dr. Richard Lucius 2. Prof. Dr. Matthias Selbach 3. Dr. Markus Landthaler Tag der mündlichen Prüfung: 21.09.2015 for my parents “No man is an island, entire of itself.” John Donne Table of Contents Table of Contents ................................................................................................................. I Abstract ............................................................................................................................... V Zusammenfassung ............................................................................................................ VI I Introduction ................................................................................................................ 7 I.1. Protein-protein interaction ........................................................................................................ 7 I.1.1. Detection of protein-protein interactions ................................................................................. 8 I.1.2. Affinity purification and mass spectrometry -

An Investigation of the Antigastric Cancer Effect in Tumor Microenvironment of Radix Rhei Et Rhizome: a Network Pharmacology Study
Hindawi Evidence-Based Complementary and Alternative Medicine Volume 2021, Article ID 9913952, 9 pages https://doi.org/10.1155/2021/9913952 Research Article An Investigation of the Antigastric Cancer Effect in Tumor Microenvironment of Radix Rhei Et Rhizome: A Network Pharmacology Study Xinmiao Wang ,1 Guanghui Zhu ,1,2 Haoyu Yang ,1,2 Ruike Gao ,1 Zhe Wu ,1 Ying Zhang,1 Xiaoyu Zhu,1 Xiaoxiao Zhang,1 and Jie Li 1 1Guang’anmen Hospital, China Academy of Chinese Medical Sciences, Beijing 100053, China 2Beijing University of Traditional Chinese Medicine, Beijing 100029, China Correspondence should be addressed to Jie Li; [email protected] Received 27 March 2021; Revised 20 May 2021; Accepted 11 June 2021; Published 24 June 2021 Academic Editor: Yibin Feng Copyright © 2021 Xinmiao Wang et al. +is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. Tumor microenvironment (TME) takes a vital effect on the occurrence and development of cancer. Radix Rhei Et Rhizome (RRER, Da-Huang in pinyin), a classical Chinese herb, has been widely used in gastric cancer (GC) for many years in China. However, inadequate systematic studies have focused on the anti-GC effect of RRER in TME. +is study intended to uncover the mechanism of it by network pharmacology. Methods. We collected compounds and targets of RRER from traditional Chinese medicine system pharmacology database and analysis platform (TCMSP) and SwissTargetPrediction. GC targets were obtained from GeneCards. Protein- protein interaction (PPI) network and RRER-GC-target network were built by STRING and Cytoscape 3.2.1. -

Gestational Exposure to Chlordecone Promotes Transgenerational
www.nature.com/scientificreports OPEN Gestational exposure to chlordecone promotes transgenerational changes in the Received: 16 January 2018 Accepted: 27 June 2018 murine reproductive system of Published: xx xx xxxx males Aurore Gely-Pernot1, Chunxiang Hao2, Louis Legof1, Luc Multigner1, Shereen Cynthia D’Cruz1, Christine Kervarrec1, Bernard Jégou1, Sergei Tevosian3 & Fatima Smagulova1 Environmental factors can afect epigenetic events during germline reprogramming and impose distinctive transgenerational consequences onto the ofspring. In this study, we examined the transgenerational efects of chlordecone (CD), an organochlorine insecticide with well-known estrogenic properties. We exposed pregnant mice to CD from embryonic day 6.5 to 15.5 and observed a reduction in spermatogonia (SG) numbers in F3, meiotic defects in spermatocytes and decrease in spermatozoa number in the frst and third generation of male progeny. The RNA qRT-PCR expression analysis in F1 and transcriptomics analysis in F3 males using the whole testes revealed changes in the expression of genes associated with chromosome segregation, cell division and DNA repair. The expression of the master regulator of pluripotency, Pou5f1, decreased in foetal and increased in adult F1, but not in F3 adult testes. Analysis of histone H3K4me3 distribution revealed widespread changes in its occupancy in the genome of F1 and F3 generations. We established that 7.1% of altered epigenetic marks were conserved between F1 and F3 generations. The overlapping changes common to F1 and F3 include genes implicated in cell adhesion and transcription factor activities functions. Diferential peaks observed in F1 males are signifcantly enriched in predicted ESR1 binding sites, some of which we confrmed to be functional. -

Detection of Chromosomal Abnormalities Associated with Prognosis of Non Small Cell Lung Cancer
(19) TZZ Z_T (11) EP 2 494 076 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12Q 1/68 (2006.01) 22.11.2017 Bulletin 2017/47 (86) International application number: (21) Application number: 10828825.9 PCT/US2010/053953 (22) Date of filing: 25.10.2010 (87) International publication number: WO 2011/056505 (12.05.2011 Gazette 2011/19) (54) DETECTION OF CHROMOSOMAL ABNORMALITIES ASSOCIATED WITH PROGNOSIS OF NON SMALL CELL LUNG CANCER ERKENNUNG VON CHROMOSOMENANOMALIEN IN VERBINDUNG MIT DER PROGNOSE VON NICHT KLEINZELLIGEM LUNGENKREBS DÉTECTION D’ANOMALIES CHROMOSOMIQUES ASSOCIÉES AU PRONOSTIC DU CANCER BRONCHO-PULMONAIRE À GRANDES CELLULES (84) Designated Contracting States: (74) Representative: Modiano, Micaela Nadia et al AL AT BE BG CH CY CZ DE DK EE ES FI FR GB Modiano & Partners GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO Thierschstrasse 11 PL PT RO RS SE SI SK SM TR 80538 München (DE) (30) Priority: 26.10.2009 US 254955 P (56) References cited: 26.10.2009 US 254968 P WO-A1-2010/121380 US-A1- 2003 022 257 US-A1- 2006 078 885 (43) Date of publication of application: 05.09.2012 Bulletin 2012/36 • BROËT PHILIPPE ET AL: "Prediction of clinical outcome in multiple lung cancer cohorts by (60) Divisional application: integrative genomics: implications for 17190353.7 chemotherapyselection.", CANCER RESEARCH, vol. 69, no. 3, 1 February 2009 (2009-02-01), pages (73) Proprietor: Abbott Molecular Inc. 1055-1062, XP9141980, ISSN: 1538-7445 Des Plaines, IL 60018 (US) • MÜLLER K M: "[New aspects of lung tumor pathology].", VERHANDLUNGEN DER (72) Inventors: DEUTSCHEN GESELLSCHAFT FÜR • PESTOVA, Ekaterina PATHOLOGIE 1999, vol. -

Normalization and Association Testing for Single-Cell CRISPR
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.12.439500; this version posted April 12, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Normalisr: normalization and association testing for single-cell CRISPR 2 screen and co-expression 3 Lingfei Wang 4 Broad Institute of MIT and Harvard, Cambridge, MA, USA 5 Center for Computational and Integrative Biology, Department of Molecular Biology, Massachusetts 6 General Hospital and Harvard Medical School, Boston, MA, USA 7 Department of Pathology, Massachusetts General Hospital and Harvard Medical School, Boston, MA, 8 USA 9 Contact: [email protected] 10 Abstract 11 Single-cell RNA sequencing (scRNA-seq) provides unprecedented technical and statistical potential to 12 study gene regulation but is subject to technical variations and sparsity. Here we present Normalisr, a 13 linear-model-based normalization and statistical hypothesis testing framework that unifies single-cell differ- 14 ential expression, co-expression, and CRISPR scRNA-seq screen analyses. By systematically detecting and 15 removing nonlinear confounding from library size, Normalisr achieves high sensitivity, specificity, speed, and 16 generalizability across multiple scRNA-seq protocols and experimental conditions with unbiased P-value es- 17 timation. We use Normalisr to reconstruct robust gene regulatory networks from trans-effects of gRNAs in 18 large-scale CRISPRi scRNA-seq screens and gene-level co-expression networks from conventional scRNA-seq. 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.04.12.439500; this version posted April 12, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Culture, Immortalization, and Characterization of Human Meibomian Gland Epithelial Cells
Cornea Culture, Immortalization, and Characterization of Human Meibomian Gland Epithelial Cells Shaohui Liu,1,2 Mark P. Hatton,1,2,3 Payal Khandelwal,1,2 and David A. Sullivan1,2 PURPOSE. Meibomian gland epithelial cells are essential in main- acuity, provide lubrication during blinking, interfere with bac- taining the health and integrity of the ocular surface. However, terial colonization, and prevent tear overflow.1–6 Meibomian very little is known about their physiological regulation. In this gland dysfunction (MGD), in turn, leads to tear film instability study, the cellular control mechanisms were explored, first to and evaporation1–6 and is thought to be the major cause of dry establish a defined culture system for the maintenance of eye syndrome throughout the world.7 primary epithelial cells from human meibomian glands and, However, aside from its regulation by sex steroids8–13 or second, to immortalize these cells, thereby developing a pre- the negative impact of retinoic acid,14 almost nothing is known clinical model that could be used to identify factors that regu- about the physiological control of the meibomian gland in late cell activity. health or disease. This dearth of knowledge is somewhat sur- METHODS. Human meibomian glands were removed from lid prising, given that the meibomian gland is a large sebaceous segments after surgery, enzymatically digested, and dissoci- gland, and numerous articles have been published about the neural, hormonal, and secretagogue modulation of nonocular ated. Isolated epithelial cells were cultured in media with or 15–24 without serum and/or 3T3 feeder layers. To attempt immortal- sebaceous gland tissue.