Exercise 2. RNA-Seq Part 1. Prepare the Working Directory. Part 2. Index
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Parallel Processing Here at the School of Statistics
Parallel Processing here at the School of Statistics Charles J. Geyer School of Statistics University of Minnesota http://www.stat.umn.edu/~charlie/parallel/ 1 • batch processing • R package multicore • R package rlecuyer • R package snow • grid engine (CLA) • clusters (MSI) 2 Batch Processing This is really old stuff (from 1975). But not everyone knows it. If you do the following at a unix prompt nohup nice -n 19 some job & where \some job" is replaced by an actual job, then • the job will run in background (because of &). • the job will not be killed when you log out (because of nohup). • the job will have low priority (because of nice -n 19). 3 Batch Processing (cont.) For example, if foo.R is a plain text file containing R commands, then nohup nice -n 19 R CMD BATCH --vanilla foo.R & executes the commands and puts the printout in the file foo.Rout. And nohup nice -n 19 R CMD BATCH --no-restore foo.R & executes the commands, puts the printout in the file foo.Rout, and saves all created R objects in the file .RData. 4 Batch Processing (cont.) nohup nice -n 19 R CMD BATCH foo.R & is a really bad idea! It reads in all the objects in the file .RData (if one is present) at the beginning. So you have no idea whether the results are reproducible. Always use --vanilla or --no-restore except when debugging. 5 Batch Processing (cont.) This idiom has nothing to do with R. If foo is a compiled C or C++ or Fortran main program that doesn't have command line arguments (or a shell, Perl, Python, or Ruby script), then nohup nice -n 19 foo & runs it. -

Introduction to Unix Part I: the Essentials
Introduction to Unix Part I: The Essentials Frederick J Tan Bioinformatics Research Faculty Carnegie Institution of Washington, Department of Embryology 9 April 2013 Unix, Linux, Ubuntu, Oh My! 2 A Three Hour Tour Part I: The Essentials client-server model, command-line interface, navigation, viewing files, searching, finding help Part II: Special Topics account settings, multi-tasking, programming, installing programs, file systems, system administration 3 The Awesome Power of the Client-Server Model 4 A Tale of Two Interfaces Command Line Graphical Terminal.app, Unix shell Finder.app, Galaxy breadth, cutting-edge discovery, visualization 5 Running Programs Using the Command-Line Interface command-line graphical type in the name of the to run a program program and hit <ENTER> double click on an icon (e.g. bowtie2) type in options after the program, to modify how a click on check boxes, before hitting <ENTER> program operates select from pull-down menus (e.g. bowtie2 --very-sensitive) 6 The Anatomy of a Shell Prompt workshop@ubuntu:~$ The text in between the colon (:) The $ symbol indicates The symbol and the dollar sign ($) indicates that the server is ready to indicates where what what directory you are in. perform a command. you type in will appear. /home/workshop$ $ ssh [email protected] 7 Task 1: Connect to your server and start top with -i option Start VirtualBox Start Ubuntu Secure SHell $ ssh [email protected] <ENTER> <SPACE> Shell Prompt /home/workshop$ <TAB> Start top -i $ top -i <CTRL> <UP> <DOWN> 8 Task 2: Figure -
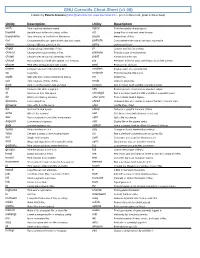
GNU Coreutils Cheat Sheet (V1.00) Created by Peteris Krumins ([email protected], -- Good Coders Code, Great Coders Reuse)
GNU Coreutils Cheat Sheet (v1.00) Created by Peteris Krumins ([email protected], www.catonmat.net -- good coders code, great coders reuse) Utility Description Utility Description arch Print machine hardware name nproc Print the number of processors base64 Base64 encode/decode strings or files od Dump files in octal and other formats basename Strip directory and suffix from file names paste Merge lines of files cat Concatenate files and print on the standard output pathchk Check whether file names are valid or portable chcon Change SELinux context of file pinky Lightweight finger chgrp Change group ownership of files pr Convert text files for printing chmod Change permission modes of files printenv Print all or part of environment chown Change user and group ownership of files printf Format and print data chroot Run command or shell with special root directory ptx Permuted index for GNU, with keywords in their context cksum Print CRC checksum and byte counts pwd Print current directory comm Compare two sorted files line by line readlink Display value of a symbolic link cp Copy files realpath Print the resolved file name csplit Split a file into context-determined pieces rm Delete files cut Remove parts of lines of files rmdir Remove directories date Print or set the system date and time runcon Run command with specified security context dd Convert a file while copying it seq Print sequence of numbers to standard output df Summarize free disk space setuidgid Run a command with the UID and GID of a specified user dir Briefly list directory -

Tour of the Terminal: Using Unix Or Mac OS X Command-Line
Tour of the Terminal: Using Unix or Mac OS X Command-Line hostabc.princeton.edu% date Mon May 5 09:30:00 EDT 2014 hostabc.princeton.edu% who | wc –l 12 hostabc.princeton.edu% Dawn Koffman Office of Population Research Princeton University May 2014 Tour of the Terminal: Using Unix or Mac OS X Command Line • Introduction • Files • Directories • Commands • Shell Programs • Stream Editor: sed 2 Introduction • Operating Systems • Command-Line Interface • Shell • Unix Philosophy • Command Execution Cycle • Command History 3 Command-Line Interface user operating system computer (human ) (software) (hardware) command- programs kernel line (text (manages interface editors, computing compilers, resources: commands - memory for working - hard-drive cpu with file - time) memory system, point-and- hard-drive many other click (gui) utilites) interface 4 Comparison command-line interface point-and-click interface - may have steeper learning curve, - may be more intuitive, BUT provides constructs that can BUT can also be much more make many tasks very easy human-manual-labor intensive - scales up very well when - often does not scale up well when have lots of: have lots of: data data programs programs tasks to accomplish tasks to accomplish 5 Shell Command-line interface provided by Unix and Mac OS X is called a shell a shell: - prompts user for commands - interprets user commands - passes them onto the rest of the operating system which is hidden from the user How do you access a shell ? - if you have an account on a machine running Unix or Linux , just log in. A default shell will be running. - if you are using a Mac, run the Terminal app. -

Gnu Coreutils Core GNU Utilities for Version 5.93, 2 November 2005
gnu Coreutils Core GNU utilities for version 5.93, 2 November 2005 David MacKenzie et al. This manual documents version 5.93 of the gnu core utilities, including the standard pro- grams for text and file manipulation. Copyright c 1994, 1995, 1996, 2000, 2001, 2002, 2003, 2004, 2005 Free Software Foundation, Inc. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.1 or any later version published by the Free Software Foundation; with no Invariant Sections, with no Front-Cover Texts, and with no Back-Cover Texts. A copy of the license is included in the section entitled “GNU Free Documentation License”. Chapter 1: Introduction 1 1 Introduction This manual is a work in progress: many sections make no attempt to explain basic concepts in a way suitable for novices. Thus, if you are interested, please get involved in improving this manual. The entire gnu community will benefit. The gnu utilities documented here are mostly compatible with the POSIX standard. Please report bugs to [email protected]. Remember to include the version number, machine architecture, input files, and any other information needed to reproduce the bug: your input, what you expected, what you got, and why it is wrong. Diffs are welcome, but please include a description of the problem as well, since this is sometimes difficult to infer. See section “Bugs” in Using and Porting GNU CC. This manual was originally derived from the Unix man pages in the distributions, which were written by David MacKenzie and updated by Jim Meyering. -

Unix Scripts and Job Scheduling
UnixUnix ScriptsScripts andand JobJob SchedulingScheduling Michael B. Spring Department of Information Science and Telecommunications University of Pittsburgh [email protected] http://www.sis.pitt.edu/~spring OverviewOverview Ü Shell Scripts Shell script basics Variables in shell scripts Korn shell arithmetic Commands for scripts Flow control, tests, and expressions Making Scripts Friendlier Functions Pipes and Shell Scripts Scripts with awk and/or sed Ü Job Scheduling bg and at cron RunningRunning aa ShellShell ScriptScript Ü First three forms spawn a new process, so new variable values are not left when you return sh < filename – where sh is the name of a shell – does not allow arguments sh filename filename – Assumes directory in path – Assumes chmod +x filename . filename – Does not spawn a new shell. – Changes to system variables impact the current shell Ü you may exit a shell script by Getting to the last line Encountering an exit command Executing a command that results in an error condition that causes an exit. StructureStructure ofof aa ShellShell ScriptScript Ü Basic structure #! Program to execute script # comment Commands and structures Ü Line continuation | at the end of the line is an assumed continuation \ at the end of a line is an explicit continuation Ü # in a shell script indicates a comment to \n Ü Back quotes in command cause immediate execution and substitution DebuggingDebugging aa scriptscript Ü Use the command set –x within a script Ü You can also activate the following set options -n read commands before executing them – for testing scripts -u make it an error to reference a non existing file -v print input as it is read - disable the –x and –v commands Ü Set the variable PS4 to some value that will help – e.g. -

Linux-Cheat-Sheet-Sponsored-By-Loggly.Pdf
Linux Command Cheat Sheet Share This Cheat Sheet Basic commands File management File Utilities Memory & Processes | Pipe (redirect) output find search for a file tr -d translate or delete character free -m display free and used system memory sudo [command] run < command> in superuser ls -a -C -h list content of directory uniq -c -u report or omit repeated lines mode killall stop all process by name rm -r -f remove files and directory split -l split file into pieces nohup [command] run < command> immune to sensors CPU temperature hangup signal locate -i find file, using updatedb(8) wc -w print newline, word, and byte database counts for each file top display current processes, real man [command] display help pages of time monitoring < command> cp -a -R -i copy files or directory head -n output the first part of files kill -1 -9 send signal to process [command] & run < command> and send task du -s disk usage cut -s remove section from file to background service manage or run sysV init script file -b -i identify the file type diff -q file compare, line by line [start|stop|restart] >> [fileA] append to fileA, preserving existing contents mv -f -i move files or directory join -i join lines of two files on a ps aux display current processes, common field snapshot > [fileA] output to fileA, overwriting grep, egrep, fgrep -i -v print lines matching pattern contents more, less view file content, one page at a dmesg -k display system messages time echo -n display a line of text sort -n sort lines in text file xargs build command line from File compression previous output -

Doing More in UNIX: Command-Line Tools HORT 59000 Lecture 3 Instructor: Kranthi Varala UNIX Features
Doing more in UNIX: Command-line tools HORT 59000 Lecture 3 Instructor: Kranthi Varala UNIX features • Command-line based. • Supports thousands of small programs running simultaneously. • Easy to create pipelines from individual programs. • Each command has 3 Input/Output streams: STDIN, STDOUT, STDERR. • Be aware of your location in the file system and permissions. Using a compute node interactively 1. ssh <yourID>@scholar.rcac.purdue.edu Log in to the head node. 2. qsub -I -l nodes=1:ppn=2,walltime=02:00:00 Log in to the compute node to run jobs interactively. Controlling processes • Foreground: Default mode for running commands. The shell waits on the process to finish. • Process retains control of the command line. • Key input is directed to the active process. • Background: Process is initiated and pushed to the background. • Control of command-line is returned to the user. • Key input and other interactions are no longer passed to the process. • Processes can be pushed to background at initiation using & • E.g., cat North_of_Boston.txt & Stopping a process • Active processes can be stopped or terminated (killed) using the SIGSTOP and SIGKILL signals. • SIGSTOP in UNIX shell is issued by ctrl+z • Once a process receives SIGSTOP it is suspended and the job number [j] is shown. • Suspended processes can pushed to background using the command bg %j . Killing a process • Active processes can be stopped or terminated (killed) using the SIGSTOP and SIGKILL signals. • SIGKILL in UNIX shell is given by ctrl+c • SIGKILL kills the process immediately and returns control to the user. • Stopped processes can be killed using kill command. -

Shell Programming in Unix, Linux and OS X: the Fourth Edition of Unix
ptg18186768 ptg18186768 An iconic symbol of the American West, Monument Valley is one of the natural wonders of the world. The red-sand desert region is located within the range of the Navajo Nation on the Arizona-Utah border and is host to towering sandstone rock formations that have been sculpted over time and soar 400 to 1,000 feet above the valley floor. Three of the valley’s most photographed peaks are the distinctive East and West Mitten Buttes and Merrick Butte. Developer’s Library ESSENTIAL REFERENCES FOR PROGRAMMING PROFESSIONALS Developer’s Library books are designed to provide practicing programmers with unique, high-quality references and tutorials on the programming languages and technologies they use in their daily work. All books in the Developer’s Library are written by expert technology practitioners who are especially skilled at organizing and presenting information in a way that’s useful for other programmers. Key titles include some of the best, most widely acclaimed books within their topic areas: PHP & MySQL Web Development Python Essential Reference Luke Welling & Laura Thomson David Beazley ISBN-13: 978-0-321-83389-1 ISBN-13: 978-0-672-32862-6 MySQL Programming in Objective-C ptg18186768 Paul DuBois Stephen G. Kochan ISBN-13: 978-0-672-32938-8 ISBN-13: 978-0-321-56615-7 Linux Kernel Development Programming in C Robert Love Stephen G. Kochan ISBN-13: 978-0-672-32946-3 ISBN-13: 978-0-321-77641-9 Developer’s Library books are available at most retail and online bookstores, as well as by subscription from Safari Books Online at safari.informit.com Developer’s Library informit.com/devlibrary Shell Programming in Unix, Linux and OS X ptg18186768 Fourth Edition Stephen G. -

Unix Shell 2
1 Unix Shell 2 Lab Objective: Introduce system management, calling Unix Shell commands within Python, and other advanced topics. As in the last lab, the majority of learning will not be had in nishing the problems, but in following the examples. File Security To begin, run the following command while inside the Shell2/Python/ directory (Shell2/ is the end product of Shell1/ from the previous lab). Notice your output will dier from that printed below; this is for learning purposes. $ ls -l -rw-rw-r-- 1 username groupname 194 Aug 5 20:20 calc.py -rw-rw-r-- 1 username groupname 373 Aug 5 21:16 count_files.py -rwxr-xr-x 1 username groupname 27 Aug 5 20:22 mult.py -rw-rw-r-- 1 username groupname 721 Aug 5 20:23 project.py Notice the rst column of the output. The rst character denotes the type of the item whether it be a normal le, a directory, a symbolic link, etc. The remaining nine characters denote the permissions associated with that le. Specically, these permissions deal with reading, wrtiting, and executing les. There are three categories of people associated with permissions. These are the user (the owner), group, and others. For example, look at the output for mult.py. The rst character - denotes that mult.py is a normal le. The next three characters, rwx tell us the owner can read, write, and execute the le. The next three characters r-x tell us members of the same group can read and execute the le. The nal three characters --x tell us other users can execute the le and nothing more. -

Linux (BASH) Cheat Sheet
freeworld.posterous.com Linux Bash Shell Cheat Sheet (works with about every distribution, except for apt-get which is Ubuntu/Debian exclusive) Legend: Everything in “<>” is to be replaced, ex: <fileName> --> iLovePeanuts.txt Don't include the '=' in your commands '..' means that more than one file can be affected with only one command ex: rm file.txt file2.txt movie.mov .. .. Linux Bash Shell Cheat Sheet Basic Commands Basic Terminal Shortcuts Basic file manipulation CTRL L = Clear the terminal cat <fileName> = show content of file CTRL D = Logout (less, more) SHIFT Page Up/Down = Go up/down the terminal head = from the top CTRL A = Cursor to start of line -n <#oflines> <fileName> CTRL E = Cursor the end of line CTRL U = Delete left of the cursor tail = from the bottom CTRL K = Delete right of the cursor -n <#oflines> <fileName> CTRL W = Delete word on the left CTRL Y = Paste (after CTRL U,K or W) mkdir = create new folder TAB = auto completion of file or command mkdir myStuff .. CTRL R = reverse search history mkdir myStuff/pictures/ .. !! = repeat last command CTRL Z = stops the current command (resume with fg in foreground or bg in background) cp image.jpg newimage.jpg = copy and rename a file Basic Terminal Navigation cp image.jpg <folderName>/ = copy to folder cp image.jpg folder/sameImageNewName.jpg ls -a = list all files and folders cp -R stuff otherStuff = copy and rename a folder ls <folderName> = list files in folder cp *.txt stuff/ = copy all of *<file type> to folder ls -lh = Detailed list, Human readable ls -l *.jpg = list jpeg files only mv file.txt Documents/ = move file to a folder ls -lh <fileName> = Result for file only mv <folderName> <folderName2> = move folder in folder mv filename.txt filename2.txt = rename file cd <folderName> = change directory mv <fileName> stuff/newfileName if folder name has spaces use “ “ mv <folderName>/ . -

GPL-3-Free Replacements of Coreutils 1 Contents
GPL-3-free replacements of coreutils 1 Contents 2 Coreutils GPLv2 2 3 Alternatives 3 4 uutils-coreutils ............................... 3 5 BSDutils ................................... 4 6 Busybox ................................... 5 7 Nbase .................................... 5 8 FreeBSD ................................... 6 9 Sbase and Ubase .............................. 6 10 Heirloom .................................. 7 11 Replacement: uutils-coreutils 7 12 Testing 9 13 Initial test and results 9 14 Migration 10 15 Due to the nature of Apertis and its target markets there are licensing terms that 1 16 are problematic and that forces the project to look for alternatives packages. 17 The coreutils package is good example of this situation as its license changed 18 to GPLv3 and as result Apertis cannot provide it in the target repositories and 19 images. The current solution of shipping an old version which precedes the 20 license change is not tenable in the long term, as there are no upgrades with 21 bugfixes or new features for such important package. 22 This situation leads to the search for a drop-in replacement of coreutils, which 23 need to provide compatibility with the standard GNU coreutils packages. The 24 reason behind is that many other packages rely on the tools it provides, and 25 failing to do that would lead to hard to debug failures and many custom patches 26 spread all over the archive. In this regard the strict requirement is to support 27 the features needed to boot a target image with ideally no changes in other 28 components. The features currently available in our coreutils-gplv2 fork are a 29 good approximation. 30 Besides these specific requirements, the are general ones common to any Open 31 Source Project, such as maturity and reliability.