Spiral Waves in Cartesian, Polar and Spherical Geometries
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Brief Information on the Surfaces Not Included in the Basic Content of the Encyclopedia
Brief Information on the Surfaces Not Included in the Basic Content of the Encyclopedia Brief information on some classes of the surfaces which cylinders, cones and ortoid ruled surfaces with a constant were not picked out into the special section in the encyclo- distribution parameter possess this property. Other properties pedia is presented at the part “Surfaces”, where rather known of these surfaces are considered as well. groups of the surfaces are given. It is known, that the Plücker conoid carries two-para- At this section, the less known surfaces are noted. For metrical family of ellipses. The straight lines, perpendicular some reason or other, the authors could not look through to the planes of these ellipses and passing through their some primary sources and that is why these surfaces were centers, form the right congruence which is an algebraic not included in the basic contents of the encyclopedia. In the congruence of the4th order of the 2nd class. This congru- basis contents of the book, the authors did not include the ence attracted attention of D. Palman [8] who studied its surfaces that are very interesting with mathematical point of properties. Taking into account, that on the Plücker conoid, view but having pure cognitive interest and imagined with ∞2 of conic cross-sections are disposed, O. Bottema [9] difficultly in real engineering and architectural structures. examined the congruence of the normals to the planes of Non-orientable surfaces may be represented as kinematics these conic cross-sections passed through their centers and surfaces with ruled or curvilinear generatrixes and may be prescribed a number of the properties of a congruence of given on a picture. -

Linguistic Presentation of Objects
Zygmunt Ryznar dr emeritus Cracow Poland [email protected] Linguistic presentation of objects (types,structures,relations) Abstract Paper presents phrases for object specification in terms of structure, relations and dynamics. An applied notation provides a very concise (less words more contents) description of object. Keywords object relations, role, object type, object dynamics, geometric presentation Introduction This paper is based on OSL (Object Specification Language) [3] dedicated to present the various structures of objects, their relations and dynamics (events, actions and processes). Jay W.Forrester author of fundamental work "Industrial Dynamics”[4] considers the importance of relations: “the structure of interconnections and the interactions are often far more important than the parts of system”. Notation <!...> comment < > container of (phrase,name...) ≡> link to something external (outside of definition)) <def > </def> start-end of definition <spec> </spec> start-end of specification <beg> <end> start-end of section [..] or {..} 1 list of assigned keywords :[ or :{ structure ^<name> optional item (..) list of items xxxx(..) name of list = value assignment @ mark of special attribute,feature,property @dark unknown, to obtain, to discover :: belongs to : equivalent name (e.g.shortname) # number of |name| executive/operational object ppppXxxx name of item Xxxx with prefix ‘pppp ‘ XXXX basic object UUUU.xxxx xxxx object belonged to UUUU object class & / conjunctions ‘and’ ‘or’ 1 If using Latex editor we suggest [..] brackets -

The Ordered Distribution of Natural Numbers on the Square Root Spiral
The Ordered Distribution of Natural Numbers on the Square Root Spiral - Harry K. Hahn - Ludwig-Erhard-Str. 10 D-76275 Et Germanytlingen, Germany ------------------------------ mathematical analysis by - Kay Schoenberger - Humboldt-University Berlin ----------------------------- 20. June 2007 Abstract : Natural numbers divisible by the same prime factor lie on defined spiral graphs which are running through the “Square Root Spiral“ ( also named as “Spiral of Theodorus” or “Wurzel Spirale“ or “Einstein Spiral” ). Prime Numbers also clearly accumulate on such spiral graphs. And the square numbers 4, 9, 16, 25, 36 … form a highly three-symmetrical system of three spiral graphs, which divide the square-root-spiral into three equal areas. A mathematical analysis shows that these spiral graphs are defined by quadratic polynomials. The Square Root Spiral is a geometrical structure which is based on the three basic constants: 1, sqrt2 and π (pi) , and the continuous application of the Pythagorean Theorem of the right angled triangle. Fibonacci number sequences also play a part in the structure of the Square Root Spiral. Fibonacci Numbers divide the Square Root Spiral into areas and angle sectors with constant proportions. These proportions are linked to the “golden mean” ( golden section ), which behaves as a self-avoiding-walk- constant in the lattice-like structure of the square root spiral. Contents of the general section Page 1 Introduction to the Square Root Spiral 2 2 Mathematical description of the Square Root Spiral 4 3 The distribution -

Semiflexible Polymers
Semiflexible Polymers: Fundamental Theory and Applications in DNA Packaging Thesis by Andrew James Spakowitz In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy California Institute of Technology MC 210-41 Caltech Pasadena, California 91125 2004 (Defended September 28, 2004) ii c 2004 Andrew James Spakowitz All Rights Reserved iii Acknowledgements Over the last five years, I have benefitted greatly from my interactions with many people in the Caltech community. I am particularly grateful to the members of my thesis committee: John Brady, Rob Phillips, Niles Pierce, and David Tirrell. Their example has served as guidance in my scientific development, and their professional assistance and coaching have proven to be invaluable in my future career plans. Our collaboration has impacted my research in many ways and will continue to be fruitful for years to come. I am grateful to my research advisor, Zhen-Gang Wang. Throughout my scientific development, Zhen-Gang’s ongoing patience and honesty have been instrumental in my scientific growth. As a researcher, his curiosity and fearlessness have been inspirational in developing my approach and philosophy. As an advisor, he serves as a model that I will turn to throughout the rest of my career. Throughout my life, my family’s love, encouragement, and understanding have been of utmost importance in my development, and I thank them for their ongoing devotion. My friends have provided the support and, at times, distraction that are necessary in maintaining my sanity; for this, I am thankful. Finally, I am thankful to my fianc´ee, Sarah Heilshorn, for her love, support, encourage- ment, honesty, critique, devotion, and wisdom. -

Coverrailway Curves Book.Cdr
RAILWAY CURVES March 2010 (Corrected & Reprinted : November 2018) INDIAN RAILWAYS INSTITUTE OF CIVIL ENGINEERING PUNE - 411 001 i ii Foreword to the corrected and updated version The book on Railway Curves was originally published in March 2010 by Shri V B Sood, the then professor, IRICEN and reprinted in September 2013. The book has been again now corrected and updated as per latest correction slips on various provisions of IRPWM and IRTMM by Shri V B Sood, Chief General Manager (Civil) IRSDC, Delhi, Shri R K Bajpai, Sr Professor, Track-2, and Shri Anil Choudhary, Sr Professor, Track, IRICEN. I hope that the book will be found useful by the field engineers involved in laying and maintenance of curves. Pune Ajay Goyal November 2018 Director IRICEN, Pune iii PREFACE In an attempt to reach out to all the railway engineers including supervisors, IRICEN has been endeavouring to bring out technical books and monograms. This book “Railway Curves” is an attempt in that direction. The earlier two books on this subject, viz. “Speed on Curves” and “Improving Running on Curves” were very well received and several editions of the same have been published. The “Railway Curves” compiles updated material of the above two publications and additional new topics on Setting out of Curves, Computer Program for Realignment of Curves, Curves with Obligatory Points and Turnouts on Curves, with several solved examples to make the book much more useful to the field and design engineer. It is hoped that all the P.way men will find this book a useful source of design, laying out, maintenance, upgradation of the railway curves and tackling various problems of general and specific nature. -

Some Curves and the Lengths of Their Arcs Amelia Carolina Sparavigna
Some Curves and the Lengths of their Arcs Amelia Carolina Sparavigna To cite this version: Amelia Carolina Sparavigna. Some Curves and the Lengths of their Arcs. 2021. hal-03236909 HAL Id: hal-03236909 https://hal.archives-ouvertes.fr/hal-03236909 Preprint submitted on 26 May 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Some Curves and the Lengths of their Arcs Amelia Carolina Sparavigna Department of Applied Science and Technology Politecnico di Torino Here we consider some problems from the Finkel's solution book, concerning the length of curves. The curves are Cissoid of Diocles, Conchoid of Nicomedes, Lemniscate of Bernoulli, Versiera of Agnesi, Limaçon, Quadratrix, Spiral of Archimedes, Reciprocal or Hyperbolic spiral, the Lituus, Logarithmic spiral, Curve of Pursuit, a curve on the cone and the Loxodrome. The Versiera will be discussed in detail and the link of its name to the Versine function. Torino, 2 May 2021, DOI: 10.5281/zenodo.4732881 Here we consider some of the problems propose in the Finkel's solution book, having the full title: A mathematical solution book containing systematic solutions of many of the most difficult problems, Taken from the Leading Authors on Arithmetic and Algebra, Many Problems and Solutions from Geometry, Trigonometry and Calculus, Many Problems and Solutions from the Leading Mathematical Journals of the United States, and Many Original Problems and Solutions. -

Spiral Pdf, Epub, Ebook
SPIRAL PDF, EPUB, EBOOK Roderick Gordon,Brian Williams | 496 pages | 01 Sep 2011 | Chicken House Ltd | 9781906427849 | English | Somerset, United Kingdom Spiral PDF Book Clear your history. Cann is on the run. Remark: a rhumb line is not a spherical spiral in this sense. The spiral has inspired artists throughout the ages. Metacritic Reviews. Fast, Simple and effective in getting high quality formative assessment in seconds. It has been nominated at the Globes de Cristal Awards four times, winning once. Name that government! This last season has two episodes less than the previous ones. The loxodrome has an infinite number of revolutions , with the separation between them decreasing as the curve approaches either of the poles, unlike an Archimedean spiral which maintains uniform line-spacing regardless of radius. Ali Tewfik Jellab changes from recurring character to main. William Schenk Christopher Tai Looking for a movie the entire family can enjoy? Looking for a movie the entire family can enjoy? Edit Cast Series cast summary: Caroline Proust June Assess in real-time or asynchronously. Time Traveler for spiral The first known use of spiral was in See more words from the same year. A hyperbolic spiral appears as image of a helix with a special central projection see diagram. TV series to watch. External Sites. That dark, messy, morally ambivalent universe they live in is recognisable even past the cultural differences, such as the astonishing blurring of the boundary between investigative police work and judgement — it's not so much uniquely French as uniquely modern. Photo Gallery. Some familiar faces, and some new characters, keep things ticking along nicely. -

Archimedean Spirals ∗
Archimedean Spirals ∗ An Archimedean Spiral is a curve defined by a polar equation of the form r = θa, with special names being given for certain values of a. For example if a = 1, so r = θ, then it is called Archimedes’ Spiral. Archimede’s Spiral For a = −1, so r = 1/θ, we get the reciprocal (or hyperbolic) spiral. Reciprocal Spiral ∗This file is from the 3D-XploreMath project. You can find it on the web by searching the name. 1 √ The case a = 1/2, so r = θ, is called the Fermat (or hyperbolic) spiral. Fermat’s Spiral √ While a = −1/2, or r = 1/ θ, it is called the Lituus. Lituus In 3D-XplorMath, you can change the parameter a by going to the menu Settings → Set Parameters, and change the value of aa. You can see an animation of Archimedean spirals where the exponent a varies gradually, from the menu Animate → Morph. 2 The reason that the parabolic spiral and the hyperbolic spiral are so named is that their equations in polar coordinates, rθ = 1 and r2 = θ, respectively resembles the equations for a hyperbola (xy = 1) and parabola (x2 = y) in rectangular coordinates. The hyperbolic spiral is also called reciprocal spiral because it is the inverse curve of Archimedes’ spiral, with inversion center at the origin. The inversion curve of any Archimedean spirals with respect to a circle as center is another Archimedean spiral, scaled by the square of the radius of the circle. This is easily seen as follows. If a point P in the plane has polar coordinates (r, θ), then under inversion in the circle of radius b centered at the origin, it gets mapped to the point P 0 with polar coordinates (b2/r, θ), so that points having polar coordinates (ta, θ) are mapped to points having polar coordinates (b2t−a, θ). -

Analysis of Spiral Curves in Traditional Cultures
Forum _______________________________________________________________ Forma, 22, 133–139, 2007 Analysis of Spiral Curves in Traditional Cultures Ryuji TAKAKI1 and Nobutaka UEDA2 1Kobe Design University, Nishi-ku, Kobe, Hyogo 651-2196, Japan 2Hiroshima-Gakuin, Nishi-ku, Hiroshima, Hiroshima 733-0875, Japan *E-mail address: [email protected] *E-mail address: [email protected] (Received November 3, 2006; Accepted August 10, 2007) Keywords: Spiral, Curvature, Logarithmic Spiral, Archimedean Spiral, Vortex Abstract. A method is proposed to characterize and classify shapes of plane spirals, and it is applied to some spiral patterns from historical monuments and vortices observed in an experiment. The method is based on a relation between the length variable along a curve and the radius of its local curvature. Examples treated in this paper seem to be classified into four types, i.e. those of the Archimedean spiral, the logarithmic spiral, the elliptic vortex and the hyperbolic spiral. 1. Introduction Spiral patterns are seen in many cultures in the world from ancient ages. They give us a strong impression and remind us of energy of the nature. Human beings have been familiar to natural phenomena and natural objects with spiral motions or spiral shapes, such as swirling water flows, swirling winds, winding stems of vines and winding snakes. It is easy to imagine that powerfulness of these phenomena and objects gave people a motivation to design spiral shapes in monuments, patterns of cloths and crafts after spiral shapes observed in their daily lives. Therefore, it would be reasonable to expect that spiral patterns in different cultures have the same geometrical properties, or at least are classified into a small number of common types. -

Superconics + Spirals 17 July 2017 1 Superconics + Spirals Cye H
Superconics + Spirals 17 July 2017 Superconics + Spirals Cye H. Waldman Copyright 2017 Introduction Many spirals are based on the simple circle, although modulated by a variable radius. We have generalized the concept by allowing the circle to be replaced by any closed curve that is topologically equivalent to it. In this note we focus on the superconics for the reasons that they are at once abundant, analytical, and aesthetically pleasing. We also demonstrate how it can be applied to random closed forms. The spirals of interest are of the general form . A partial list of these spirals is given in the table below: Spiral Remarks Logarithmic b = flair coefficient m = 1, Archimedes Archimedean m = 2, Fermat (generalized) m = -1, Hyperbolic m = -2, Lituus Parabolic Cochleoid Poinsot Genesis of the superspiral (1) The idea began when someone on line was seeking to create a 3D spiral with a racetrack planiform. Our first thought was that the circle readily transforms to an ellipse, for example Not quite a racetrack, but on the right track. The rest cascades in a hurry, as This is subject to the provisions that the random form is closed and has no crossing lines. This also brought to mind Euler’s famous derivation, connecting the five most famous symbols in mathematics. 1 Superconics + Spirals 17 July 2017 In the present paper we’re concerned primarily with the superconics because the mapping is well known and analytic. More information on superconics can be found in Waldman (2016) and submitted for publication by Waldman, Chyau, & Gray (2017). Thus, we take where is a parametric variable (not to be confused with the polar or tangential angles). -

ALL ABOUT SPIRALS a Spiral Can Be Described As Any 2D Continuous Curve Where the Radial Distance R from the Origin Equals a Specified Function of the Angle Θ
, ALL ABOUT SPIRALS A spiral can be described as any 2D continuous curve where the radial distance r from the origin equals a specified function of the angle θ. Mathematically it has the unique derivative given by- dr=(df/dθ)dθ with r=0 at θ=0 On integrating one finds- () ∫ 푑푡 r= This solution yields two simple forms. The first occurs when df/dt=α. We get- r=α(θ) This simplest of continuous spiral figures is just the Archimedes Spiral discovered by him over two thousand years ago. Setting the constant to α=3/π , we have the plot- Note that the distance between turns of the spiral remain the same. A second simple spiral is found when df/dθ=βf, where f=exp(훽휃). If we take r=1 at θ=0, it produces- r=exp(βθ) or the equivalent ln(r)=βθ It is known as the Logarithmic Spiral or Bernoulli’s Spiral. Here is its graph when β=1/10- A property of the Bernouli Spiral is that the angle between any radial line and the tangent to the spiral remains a constant. J. Bernoulli was so intrigued by this spiral that he had a copy placed on his tombstone in Basel Switzerland. Here I am pointing to it back in 2000 - One can construct numerous other spirals by simply changing the form of df(θ)/dθ and picking a starting point r(θ0)=r0. So if df/dθ=α/[2sqrt(θ)] and r=0 when θ=0, we get the spiral– r=αsqrt(θ) It looks as follows- This figure is known as the Fermat Spiral. -
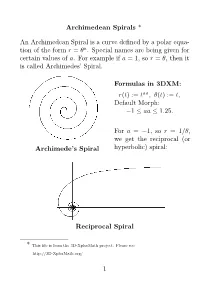
Archimedean Spirals * an Archimedean Spiral Is a Curve
Archimedean Spirals * An Archimedean Spiral is a curve defined by a polar equa- tion of the form r = θa. Special names are being given for certain values of a. For example if a = 1, so r = θ, then it is called Archimedes’ Spiral. Formulas in 3DXM: r(t) := taa, θ(t) := t, Default Morph: 1 aa 1.25. − ≤ ≤ For a = 1, so r = 1/θ, we get the− reciprocal (or Archimede’s Spiral hyperbolic) spiral: Reciprocal Spiral * This file is from the 3D-XplorMath project. Please see: http://3D-XplorMath.org/ 1 The case a = 1/2, so r = √θ, is called the Fermat (or hyperbolic) spiral. Fermat’s Spiral While a = 1/2, or r = 1/√θ, it is called the Lituus: − Lituus 2 In 3D-XplorMath, you can change the parameter a by go- ing to the menu Settings Set Parameters, and change the value of aa. You can see→an animation of Archimedean spirals where the exponent a = aa varies gradually, be- tween 1 and 1.25. See the Animate Menu, entry Morph. − The reason that the parabolic spiral and the hyperbolic spiral are so named is that their equations in polar coor- dinates, rθ = 1 and r2 = θ, respectively resembles the equations for a hyperbola (xy = 1) and parabola (x2 = y) in rectangular coordinates. The hyperbolic spiral is also called reciprocal spiral be- cause it is the inverse curve of Archimedes’ spiral, with inversion center at the origin. The inversion curve of any Archimedean spirals with re- spect to a circle as center is another Archimedean spiral, scaled by the square of the radius of the circle.