Impact of Hybridisation in Two [I]Cossypha[I] Robin-Chat Species In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Flyer200206 Parent
THE, FLYE, R Volume 26,25, NumberNumber 6 6 tuneJune 20022002 NEXT MEETING After scientists declared the Gunnison Sage Grouse a new species two years ago, a wide spot on Gun- Summer is here and we won't have meeting until a nison County (Colorado) Road 887, has become an Wednesday, 18. It begin September will atl:30 international bird-watching sensation. Birders from p.m. in Room 117 Millington Hall, on the William around the ,world wait silently in the cold dark of a campus. The editors'also get a summer &Mary Colorado spring pre-dawn to hear the "Thwoomp! vacation so there will be no July Flyer, but "God Thwoomp! Thwoomp!" of a male Gunnison Sage the creek rise," there be an willing and if don't will Grouse preparing to mate. The noise comes from August issue. specialized air sacs on the bird's chest. And this is now one stop on a well-traveled 1,000 mile circuit being traveled by birders wanting to add this Gun- RAIN CURTAILS FIELD TRIP TO nison bird, plus the Chukar, the Greater Sage YORK RIVER STATE PARK Grouse, the White-tailed Ptarmigan, the Greater Chicken Skies were threatening and the wind was fierce at Prairie and the Lesser Prairie Chicken to the beginning of the trip to the York River State their lii'e lists. Park on May 18. Despite all of that, leader Tom It was not always like this. Prior to the two-year- Armour found some very nice birds before the rains ago decision by the Ornithological Union that this came flooding down. -

Rare Birds of California Now Available! Price $54.00 for WFO Members, $59.99 for Nonmembers
Volume 40, Number 3, 2009 The 33rd Report of the California Bird Records Committee: 2007 Records Daniel S. Singer and Scott B. Terrill .........................158 Distribution, Abundance, and Survival of Nesting American Dippers Near Juneau, Alaska Mary F. Willson, Grey W. Pendleton, and Katherine M. Hocker ........................................................191 Changes in the Winter Distribution of the Rough-legged Hawk in North America Edward R. Pandolfino and Kimberly Suedkamp Wells .....................................................210 Nesting Success of California Least Terns at the Guerrero Negro Saltworks, Baja California Sur, Mexico, 2005 Antonio Gutiérrez-Aguilar, Roberto Carmona, and Andrea Cuellar ..................................... 225 NOTES Sandwich Terns on Isla Rasa, Gulf of California, Mexico Enriqueta Velarde and Marisol Tordesillas ...............................230 Curve-billed Thrasher Reproductive Success after a Wet Winter in the Sonoran Desert of Arizona Carroll D. Littlefield ............234 First North American Records of the Rufous-tailed Robin (Luscinia sibilans) Lucas H. DeCicco, Steven C. Heinl, and David W. Sonneborn ........................................................237 Book Reviews Rich Hoyer and Alan Contreras ...........................242 Featured Photo: Juvenal Plumage of the Aztec Thrush Kurt A. Radamaker .................................................................247 Front cover photo by © Bob Lewis of Berkeley, California: Dusky Warbler (Phylloscopus fuscatus), Richmond, Contra Costa County, California, 9 October 2008, discovered by Emilie Strauss. Known in North America including Alaska from over 30 records, the Dusky is the Old World Warbler most frequent in western North America south of Alaska, with 13 records from California and 2 from Baja California. Back cover “Featured Photos” by © Kurt A. Radamaker of Fountain Hills, Arizona: Aztec Thrush (Ridgwayia pinicola), re- cently fledged juvenile, Mesa del Campanero, about 20 km west of Yecora, Sonora, Mexico, 1 September 2007. -

Does a Rival's Song Elicit Territorial Defense in a Tropical Songbird, The
ABC 2017, 4(2):146-153 Animal Behavior and Cognition https://doi.org/10.12966/abc.02.05.2017 ©Attribution 3.0 Unported (CC BY 3.0) Does a Rival’s Song Elicit Territorial Defense in a Tropical Songbird, the Pied Bush Chat (Saxicola caprata)? Navjeevan Dadwal1* and Dinesh Bhatt1 1Gurukula Kangri University, Haridwar, Uttarakhand, India *Corresponding author (Email:[email protected]) Citation – Dadwal, N., & Bhatt, D. (2017). Does a rival’s song elicit territorial defense in a tropical songbird, the Pied Bush Chat (Saxicola caprata)? Animal Behavior and Cognition, 4(2), 146–153. https://doi.org/10.12966/ abc.02.05.2017 Abstract -The purpose of bird song and the way in which it is delivered has been argued to be adapted mainly for territorial defense. We performed a field experiment with the combination of playbacks and a model to test how much song actually relates to increased territorial defense in the territorial tropical songbird, the Pied Bush Chat, during breeding season (Feb–May, 2015) at Haridwar, Himalayan Foothills, India. As expected, the results of the experiment indicated that song was the major cue used by territory holders to cope with rival intrusions. The song rate was particularly escalated during simulated territorial interactions when the model was presented with a playback song of conspecifics. Behaviors such as restlessness (perch change), the height of perch, and distance from the model appeared to be of relatively lesser importance. To our knowledge, no avian species from the Indian subcontinent has been studied to provide evidence that song can escalate aggressive response by a territory owner. -
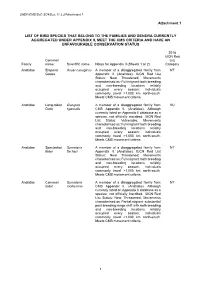
Attachment 1 LIST of BIRD SPECIES THAT BELONG to the FAMILIES
UNEP/CMS/ScC-SC4/Doc.11.3.2/Attachment 1 Attachment 1 LIST OF BIRD SPECIES THAT BELONG TO THE FAMILIES AND GENERA CURRENTLY AGGREGATED UNDER APPENDIX II, MEET THE CMS CRITERIA AND HAVE AN UNFAVOURABLE CONSERVATION STATUS 2018 IUCN Red Common List Family name Scientific name Notes for Appendix II (Sheets 1 or 2) Category Anatidae Emperor Anser canagicus A member of a disaggregated family from NT Goose Appendix II. (Anatidae). IUCN Red List Status: Near Threatened; Movements characterised as: Full migrant: both breeding and non-breeding locations reliably occupied every season; individuals commonly travel >1,000 km north-south. Meets CMS movement criteria. Anatidae Long-tailed Clangula A member of a disaggregated family from VU Duck hyemalis CMS Appendix II. (Anatidae). Although currently listed on Appendix II database as a species, not officially inscribed. IUCN Red List Status: Vulnerable; Movements characterised as: Full migrant: both breeding and non-breeding locations reliably occupied every season; individuals commonly travel >1,000 km north-south. Meets CMS movement criteria. Anatidae Spectacled Somateria A member of a disaggregated family from NT Eider fischeri Appendix II. (Anatidae). IUCN Red List Status: Near Threatened; Movements characterised as: Full migrant: both breeding and non-breeding locations reliably occupied every season; individuals commonly travel >1,000 km north-south. Meets CMS movement criteria. Anatidae Common Somateria A member of a disaggregated family from NT Eider mollissima CMS Appendix II. (Anatidae). Although currently listed on Appendix II database as a species, not officially inscribed. IUCN Red List Status: Near Threatened; Movements characterised as: Partial migrant: substantial post-breeding range shift with both breeding and non-breeding locations reliably occupied every season; individuals commonly travel >1,000 km north-south. -

Birdlife International for the Input of Analyses, Technical Information, Advice, Ideas, Research Papers, Peer Review and Comment
UNEP/CMS/ScC16/Doc.10 Annex 2b CMS Scientific Council: Flyway Working Group Reviews Review 2: Review of Current Knowledge of Bird Flyways, Principal Knowledge Gaps and Conservation Priorities Compiled by: JEFF KIRBY Just Ecology Brookend House, Old Brookend, Berkeley, Gloucestershire, GL13 9SQ, U.K. June 2010 Acknowledgements I am grateful to colleagues at BirdLife International for the input of analyses, technical information, advice, ideas, research papers, peer review and comment. Thus, I extend my gratitude to my lead contact at the BirdLife Secretariat, Ali Stattersfield, and to Tris Allinson, Jonathan Barnard, Stuart Butchart, John Croxall, Mike Evans, Lincoln Fishpool, Richard Grimmett, Vicky Jones and Ian May. In addition, John Sherwell worked enthusiastically and efficiently to provide many key publications, at short notice, and I’m grateful to him for that. I also thank the authors of, and contributors to, Kirby et al. (2008) which was a major review of the status of migratory bird species and which laid the foundations for this work. Borja Heredia, from CMS, and Taej Mundkur, from Wetlands International, also provided much helpful advice and assistance, and were instrumental in steering the work. I wish to thank Tim Jones as well (the compiler of a parallel review of CMS instruments) for his advice, comment and technical inputs; and also Simon Delany of Wetlands International. Various members of the CMS Flyway Working Group, and other representatives from CMS, BirdLife and Wetlands International networks, responded to requests for advice and comment and for this I wish to thank: Olivier Biber, Joost Brouwer, Nicola Crockford, Carlo C. Custodio, Tim Dodman, Roger Jaensch, Jelena Kralj, Angus Middleton, Narelle Montgomery, Cristina Morales, Paul Kariuki Ndang'ang'a, Paul O’Neill, Herb Raffaele and David Stroud. -

Going Beyond
Going Beyond THE BIDVEST GROUP LIMITED DIRECTORY APRIL 2018 Index and Directory DirectoryAddingTitle value Name across Telephone Location the national economy Directory 2017 The Bidvest Group Limited Bidvest directory Title Name Telephone Location BIDVEST CORPORATE Bidvest Corporate Services Group chief executive Lindsay Ralphs +27 (11) 772 8700 Johannesburg Bidvest House, 18 Crescent Drive, Chief financial officer Mark Steyn Melrose Arch Executive director Gillian McMahon PO Box 87274, Houghton, 2041 Executive director Mpumi Madisa www.bidvest.com Group company secretarial Ilze Roux Group accounting Paul Roberts Group financial manager Lorinda Petzer Group internal audit Lauren Berrington Group taxation Dharam Naidoo Group retirement funds Lesley Jurgensen +27 (10) 730 0822 Johannesburg Group Treasurer Neil Taylor Corporate finance executive Phathu Tshivhengwa Corporate affairs executive Ilze Roux Business development executive Ravi Govender Financial manager Priscilla da Rocha Business relationship manager Amanda Davidson Commercial executive Akona Ngcuka Events and marketing manager Julie Alexander Bidvest Properties Bidvest House, 18 Crescent Drive, Chief financial officer Ferdi Smit +27 (11) 772 8786 Melrose Arch, 2196 Operational director Warren Kretzmar +27 (11) 772 8725 PO Box 87274, Houghton, 2041 Legal director Colleen Krige +27 (11) 772 8729 Fax: +27 (11) 772 8970 Bidvest Procurement Managing director Derek Kinnear +27 (11) 243 2040 Johannesburg Ground Floor, 158 Jan Smuts Avenue, Financial manager Ilze Rossouw Rosebank, 2196 Operations -

Disaggregation of Bird Families Listed on Cms Appendix Ii
Convention on the Conservation of Migratory Species of Wild Animals 2nd Meeting of the Sessional Committee of the CMS Scientific Council (ScC-SC2) Bonn, Germany, 10 – 14 July 2017 UNEP/CMS/ScC-SC2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II (Prepared by the Appointed Councillors for Birds) Summary: The first meeting of the Sessional Committee of the Scientific Council identified the adoption of a new standard reference for avian taxonomy as an opportunity to disaggregate the higher-level taxa listed on Appendix II and to identify those that are considered to be migratory species and that have an unfavourable conservation status. The current paper presents an initial analysis of the higher-level disaggregation using the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World Volumes 1 and 2 taxonomy, and identifies the challenges in completing the analysis to identify all of the migratory species and the corresponding Range States. The document has been prepared by the COP Appointed Scientific Councilors for Birds. This is a supplementary paper to COP document UNEP/CMS/COP12/Doc.25.3 on Taxonomy and Nomenclature UNEP/CMS/ScC-Sc2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II 1. Through Resolution 11.19, the Conference of Parties adopted as the standard reference for bird taxonomy and nomenclature for Non-Passerine species the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World, Volume 1: Non-Passerines, by Josep del Hoyo and Nigel J. Collar (2014); 2. -

Welcome to KPMG Crescent
Jan Smuts Ave St Andrews M1 Off Ramp Winchester Rd Jan Smuts Off Ramp Welcome to KPMGM27 Crescent M1 North On Ramp De Villiers Graaff Motorway (M1) 85 Empire Road, Parktown St Andrews Rd Albany Rd GPS Coordinates Latitude: -26.18548 | Longitude: 28.045142 85 Empire Road, Johannesburg, South Africa M1 B M1 North On Ramp Directions: From Sandton/Pretoria M1 South Take M1 (South) towards Johannesburg On Ramp Jan Smuts / Take Empire off ramp, at the robot turn left to the KPMG main St Andrews gate. (NB – the Empire entrance is temporarily closed). Continue Off Ramp to Jan Smuts Avenue, turn left and then first left into entrance on Empire Jan Smuts. M1 Off Ramp From South of JohannesburgWellington Rd /M2 Sky Bridge 4th Floor Take M1 (North) towards Sandton/Pretoria Take Exit 14A for Jan Smuts Avenue toward M27 and turn right M27 into Jan Smuts. At Empire Road turn right, at first traffic lights M1 South make a U-turn and travel back on Empire, and left into Jan Smuts On Ramp M17 Jan Smuts Ave Avenue, and first left into entrance. Empire Rd KPMG Entrance KPMG Entrance temporarily closed Off ramp On ramp T: +27 (0)11 647 7111 Private Bag 9, Jan Jan Smuts Ave F: +27 (0)11 647 8000 Parkview, 2122 E m p ire Rd Welcome to KPMG Wanooka Place St Andrews Rd, Parktown NORTH GPS Coordinates Latitude: -26.182416 | Longitude: 28.03816 St Andrews Rd, Parktown, Johannesburg, South Africa M1 St Andrews Off Ramp Jan Smuts Ave Directions: Winchester Rd From Sandton/Pretoria Take M1 (South) towards Johannesburg Take St Andrews off ramp, at the robot drive straight to the KPMG Jan Smuts main gate. -

Legal Notices Wetlike Kennisgewings
Vol. 632 Pr t ri 16 February 2018 e 0 af Februane No. 41444 LEGAL NOTICES WETLIKE KENNISGEWINGS SALES IN EXECUTION AND OTHER PUBLIC SALES GEREGTELIKE EN ANDER QPENBARE VERKOPE 2 No. 41444 GOVERNMENT GAZETTE, 16 FEBRUARY 2018 IMPORTANT NOTICE: THE GOVERNMENT PRINTING WORKS WILL NOT BE HELD RESPONSIBLE FOR ANY ERRORS THAT MIGHT OCCUR DUE TO THE SUBMISSION OF INCOMPLETE / INCORRECT / ILLEGIBLE COPY. NO FUTURE QUERIES WILL BE HANDLED IN CONNECTION WITH THE ABOVE. CONTENTS / INHOUD LEGAL NOTICES / WETLIKE KENNISGEWINGS SALES IN EXECUTION AND OTHER PUBLIC SALES GEREGTELIKE EN ANDER OPENBARE VERKOPE Sales in execution • Geregtelike verkope ....................................................................................................... 11 Gauteng ...................................................................................................................................... 11 Eastern Cape / Oos-Kaap ................................................................................................................ 44 Free State / Vrystaat ....................................................................................................................... 45 KwaZulu-Natal .............................................................................................................................. 47 Limpopo ...................................................................................................................................... 50 Mpumalanga ............................................................................................................................... -

Download Download
Ornithological Observations http://oo.adu.org.za An electronic journal published by the Animal Demography Unit at the University of Cape Town and BirdLife South Africa Ornithological Observations accepts papers containing faunistic information about birds. This includes descriptions of distribution, behaviour, breeding, foraging, food, movement, measurements, habitat and plumage. It will also consider for publication a variety of other interesting or relevant ornithological material: reports of projects and conferences, annotated checklists for a site or region, specialist bibliographies, and any other interesting or relevant material. Editor: Arnold van der Westhuizen CHORISTER ROBIN-CHAT COSSYPHA DICHROA GLEANING ON BUSHBUCK TRAGELAPHUS SCRIPTUS Craig T Symes, Theresa Rees and Karin Nelson Recommended citation format: Symes CT, Rees T, Nelson K 2015. Chorister Robin-Chat Cossypha dichroa gleaning on Bushbuck Tragelaphus scriptus. Ornithological Observations, Vol 6: 44-45 URL: http://oo.adu.org.za/content.php?id=174 Published online: 07 March 2015 - ISSN 2219-0341 - Ornithological Observations, Vol 6: 44-45 44 CHORISTER ROBIN-CHAT COSSYPHA DICHROA GLEANING ON BUSHBUCK TRAGELAPHUS SCRIPTUS Craig T Symes*1, Theresa Rees2 and Karin Nelson3 1 School of Animal, Plant & Environmental Sciences, University of the Witwatersrand, Private Bag 3, Wits 2050, South Africa 2 40 Budleigh Road, Hilton, South Africa 3 12 Sakabula Drive, Merrivale, Howick 3291, South Africa * Corresponding author: [email protected] A review of African birds feeding in association with mammals, by Fig 1 - Screen-shots of the film footage of a Chorister Robin-Chat visiting a Dean and MacDonald (1981), comprehensively documents a number Bushbuck. See footage on YouTube at http://youtu.be/D87WA5zb-9A of mammal and bird-feeding interactions. -

Comprehensive Angola 2019 Tour Report
BIRDING AFRICA THE AFRICA SPECIALISTS Comprehensive Angola 2019 Tour Report Swierstra's Francolin Text by tour leader Michael Mills Photos by tour participant Bob Zook SUMMARY ESSENTIAL DETAILS With camping on Angolan bird tours now ancient history, our fourth all- hotel-accommodated bird tour of Angola was an overwhelming success Dates 16 Aug : Kalandula to N'dalatando. both for birds and comfort. Thanks to new hotels opening up and a second 12-29 August 2019 17 Aug : N'dalatando to Muxima via northern escarpment forests of Tombinga Pass. wave of road renovations almost complete, Angola now offers some of the Birding Africa Tour Report Tour Africa Birding most comfortable travel conditions on the African continent, although Leader 18 Aug : Dry forests in Muxima area. Report Tour Africa Birding 19 Aug : Muxima to Kwanza River mouth. longer drives are needed to get to certain of the birding sites. Michael Mills 20 Aug : Kwanza River to Conda. Participants 21 Aug : Central escarpment forest at Kumbira. Andrew Cockburn 22 Aug : Conda to Mount Moco region. Mike Coverdale 23 Aug : Grasslands and montane forest at Mount Moco. Daragh Croxson 24 Aug : Dambos and miombo woodlands in the Stephen Eccles Mount Moco region. Ola Sundberg 25 Aug : Margaret's Batis hike at Mount Moco. Brazza's Martin Bob Zook 26 Aug : Mount Moco to Benguela area via wetlands of Lobito. Itinerary 27 Aug : Benguela to Lubango via rocky hillsides Besides the logistics running very smoothly we fared There were many other great birds seen too, and the and arid savannas. exceptionally well on the birds, with all participants impressive diversity of habitats meant that we logged 12 Aug : Luanda to Uíge. -

Siberian Blue Robin Larvivora Cyane from the Barak Valley of Assam with a Status Update for India
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/338137338 Siberian Blue Robin Larvivora cyane from the Barak Valley of Assam with a status update for India Article in Indian BIRDS · December 2019 CITATIONS READS 0 7 2 authors: Rejoice Gassah Vijay Anand Ismavel Makunda Christian Hospital 4 PUBLICATIONS 0 CITATIONS 22 PUBLICATIONS 12 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: Neglected health problems in rural India View project Biodiversity Documentation View project All content following this page was uploaded by Vijay Anand Ismavel on 24 December 2019. The user has requested enhancement of the downloaded file. Correspondence 123 peninsular India (Grimmett et al. 2011; Rasmussen & Anderton minY=11.103753465762485&env.maxX=93.01342361450202&env.maxY=12.31963 2012; eBird 2019). This species is a common winter visitor to 3103994705&zh=true&gp=true&ev=Z&mr=on&bmo=1&emo=1&yr=cur&byr=2019 Myanmar, Thailand, Laos, and Tonkin (Robson 2008). &eyr=2019. [Accessed on: 19 January 2019.] DeCandido, R., Subedi, T., Siponen, M., Sutasha, K., Pierce, A., Nualsri, C., & Round, P. D., 2013. Flight identification of Milvus migrans lineatus ‘Black-eared’ Kite and Milvus migrans govinda ‘Pariah’ Kite in Nepal and Thailand. BirdingASIA 20: 32–36. Grimmett, R., Inskipp, C., & Inskipp, T., 2011. Birds of the Indian Subcontinent. 2nd ed. London: Oxford University Press & Christopher Helm. Pp. 1–528. Rasmussen, P. C., & Anderton, J. C., 2012. Birds of South Asia: the Ripley guide: field guide. 2nd ed. Washington, D.C.