Global Strategy for the Ex Situ Conservation and Use of Barley Germplasm
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
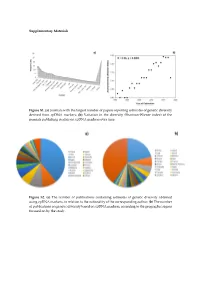
(A) Journals with the Largest Number of Papers Reporting Estimates Of
Supplementary Materials Figure S1. (a) Journals with the largest number of papers reporting estimates of genetic diversity derived from cpDNA markers; (b) Variation in the diversity (Shannon-Wiener index) of the journals publishing studies on cpDNA markers over time. Figure S2. (a) The number of publications containing estimates of genetic diversity obtained using cpDNA markers, in relation to the nationality of the corresponding author; (b) The number of publications on genetic diversity based on cpDNA markers, according to the geographic region focused on by the study. Figure S3. Classification of the angiosperm species investigated in the papers that analyzed genetic diversity using cpDNA markers: (a) Life mode; (b) Habitat specialization; (c) Geographic distribution; (d) Reproductive cycle; (e) Type of flower, and (f) Type of pollinator. Table S1. Plant species identified in the publications containing estimates of genetic diversity obtained from the use of cpDNA sequences as molecular markers. Group Family Species Algae Gigartinaceae Mazzaella laminarioides Angiospermae Typhaceae Typha laxmannii Angiospermae Typhaceae Typha orientalis Angiospermae Typhaceae Typha angustifolia Angiospermae Typhaceae Typha latifolia Angiospermae Araliaceae Eleutherococcus sessiliflowerus Angiospermae Polygonaceae Atraphaxis bracteata Angiospermae Plumbaginaceae Armeria pungens Angiospermae Aristolochiaceae Aristolochia kaempferi Angiospermae Polygonaceae Atraphaxis compacta Angiospermae Apocynaceae Lagochilus macrodontus Angiospermae Polygonaceae Atraphaxis -

An Unmöglichen Dingen Soll Man Selten Verzweifeln, an Schweren Nie
An unmöglichen Dingen soll man selten verzweifeln, an schweren nie. Johann Wolfgang von Goethe Cytogenetic mapping of BAC contigs assigned to barley chromosome 3H and comparative subchromosomal analysis within the genus Hordeum Kumulative Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften doctor rerum naturalium (Dr. rer. nat) der Naturwissenschaftlichen Fakultät III- Agrar- und Ernährungswissenschaften, Geowissenschaften und Informatik der Martin-Luther-Universität Halle-Wittenberg Vorgelegt von Lala Aliyeva-Schnorr (geb. Aliyeva) geboren am 04.04.1985 in Baku, Aserbaidschan 1. Gutachter: Prof. Dr. Klaus Pillen 2. Gutachter: Prof. Dr. Thomas Schmidt 3. Gutachter: Dr. habil. Andreas Houben Verteidigung am 21. November 2016 Acknowledgements Acknowledgements This work was performed from April 2012 till February 2016 at the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben. The financial support from IPK made the start of this project possible. Since September 2013 it was funded by German Research Foundation (DFG – Deutsche Forschungsgemeinschaft). First of all I would like to express my special thanks to Dr. habil. Andreas Houben, the head of Chromosome Structure and Function (CSF) group, for giving me the opportunity to be the part of his team, for continuous guidance, permanent encouragement as well as fruitful discussions. Also, I would like to thank Dr. Nils Stein for his help, support and discussions during my time at IPK. Special thanks to Prof. Klaus Pillen for his supportive supervision of my doctoral research and to Prof. Thomas Schmidt for the review of this thesis. My deepest thanks to all present and former members as well as visitors of the group CSF. All of them made our group an enjoyable place to work. -

Appendix C Plant and Animal Species Observed
Appendix C Plant and Animal Species Observed This list includes vascular plants, mammals, birds, reptiles, and amphibians observed in the BSA by biologists during various surveys in 2005 and 2006. This list does not include invertebrate species. Invertebrates that would be most commonly encountered on the site would include butterflies, flies, dragonflies, damselflies, beetles, earwigs, grasshoppers, crickets, termites, true bugs, mantids, lacewings, bees, wasps, ants, and spiders. PLANT SPECIES OBSERVED Scientific Name Common Name Invasive Plant Rating CLUB AND SPIKE MOSSES Selaginellaceae Spike moss family Selaginella cinerascens Mesa spikemoss TRUE FERNS Azollaceae Mosquito fern family Azolla filiculoides Pacific mosquito fern Marsileaceae Marsilea family Marsilea vestita Hairy waterclover Pilularia americana American pillwort Pteridaceae Lip fern family Pellaea andromedifolia Coffee fern Pentagramma triangularis Goldenback fern PINOPHYTA GYMNOSPERMS Cupressaceace Cypress family Juniperus californica California juniper DICOT FLOWERING PLANTS Aizoaceae Carpet weed family Carpobrotus edulis* Hottentot-fig HIGH Mesembryanthemum nodiflorum* Slender-leaved ice plant Trianthema portulacastrum Horse-purslane Amaranthaceae Amaranth family Amaranthus albus* Tumbling pigweed Amaranthus palmeri Palmer’s pigweed Amaranthus sp. Pigweed Anacardiaceae Sumac family Malosma laurina Laurel sumac Rhus ovata Sugar bush Rhus trilobata Skunkbush sumac Schinus molle* Peruvian pepper tree LIMITED Schinus terebinthifolius* Brazilian pepper tree LIMITED Mid -

Botanic Gardens Are Important Contributors to Crop Wild Relative Preservation
Published November 21, 2019 RESEARCH Botanic Gardens Are Important Contributors to Crop Wild Relative Preservation Abby Meyer* and Nicholas Barton Botanic Gardens Conservation International US at at The Huntington ABSTRACT Library, Art Museum, and Botanical Gardens, 1151 Oxford Rd., San Humans rely on crop wild relatives (CWRs) for Marino, CA 91108. Received 2 June 2019. Accepted 11 Oct. 2019. sustainable agriculture and food security through *Corresponding author ([email protected]). Assigned to Associate augmentation of crop yield, disease resistance, Editor Joseph Robins. and climatic tolerance, among other important Abbreviations: BGCI, Botanic Gardens Conservation International; traits. Many CWRs are underrepresented in crop CWR, crop wild relative; GRIN, Germplasm Resources Information gene banks. With at least one-third of known Network. plant species maintained in botanic garden living collections, the botanic garden community serves as an important global ex situ network rop wild relatives (CWRs), plants that have “an indirect that supports plant conservation and research Cuse derived from its relatively close genetic relationship to a around the world. We sought to characterize crop,” provide important genetic diversity needed by breeders and botanic garden holdings of CWRs and demon- scientists to develop a wide range of crop plant adaptations (Maxted strate capacity for cross-sector coordination in et al., 2006, p. 2680). Benefits such as increased production, better support of CWR ex situ preservation. To do this, nutrition, drought tolerance, and pest and disease resistance have Botanic Gardens Conservation International been made possible through the use of CWRs and allowed for US (BGCI-US), in partnership with the United more consistent and sustainable yields of conventional crops for States Botanic Garden, used the BGCI Plant- Search database to conduct an ex situ survey of decades (Dempewolf et al., 2017; Guarino and Lobell, 2011; Hajjar CWRs maintained in botanic gardens. -

3 November 2017 Reports
August 2017 IT/GB-7/17/24 E Item 17.4 of the Provisional Agenda SEVENTH SESSION OF THE GOVERNING BODY Kigali, Rwanda, 30 October – 3 November 2017 Reports from Institutions that have Concluded Agreements with the Governing Body under Article 15 of the International Treaty Executive Summary The document contains the Reports provided by the Centres of the Consultative Group on International Agricultural Research and one other International Institution that have concluded agreements with the Governing Body pursuant to Article 15 of the International Treaty, for the information and consideration of the Governing Body. The reports are provided, as they were received by the Secretariat, in the Appendices to this document. The document also provides update on activities carried out by the Secretariat and some recent developments, during the biennium, in regard to international germplasm collections that are held under Article 15 of the Treaty, whose continued maintenance has been reported to be experiencing major difficulties or are under threat. Guidance Sought The Governing Body is invited to consider the reports and the information provided in this document, and provide further guidance, taking into account the elements for a possible Resolution provided in Appendix 1 to this Document. This document can be accessed using the Quick Response Code on this page; an FAO initiative to minimize its environmental impact and promote greener communications. Other documents can be consulted at http://www.fao.org/plant-treaty/meetings/meetings- detail/en/c/888771/ 2 IT/GB-7/17/24 Table of Contents Paragraphs I. INTRODUCTION ........................................................................................................ 1 – 2 II. SUMMARY OF THE REPORTS RECEIVED ....................................................... -

Structural Characteristics of Scbx Genes Controlling the Biosynthesis of Hydroxamic Acids in Rye (Secale Cereale L.)
J Appl Genetics (2015) 56:287–298 DOI 10.1007/s13353-015-0271-z PLANT GENETICS • ORIGINAL PAPER Structural characteristics of ScBx genes controlling the biosynthesis of hydroxamic acids in rye (Secale cereale L.) Beata Bakera & Bogna Makowska & Jolanta Groszyk & Michał Niziołek & Wacław Orczyk & Hanna Bolibok-Brągoszewska & Aneta Hromada-Judycka & Monika Rakoczy-Trojanowska Received: 15 July 2014 /Revised: 12 January 2015 /Accepted: 13 January 2015 /Published online: 10 February 2015 # The Author(s) 2015. This article is published with open access at Springerlink.com Abstract Benzoxazinoids (BX) are major secondary metab- analyses of the resulting contigs were used to examine the olites of gramineous plants that play an important role in dis- structure and other features of these genes, including their ease resistance and allelopathy. They also have many other promoters, introns and 3’UTRs. Comparative analysis unique properties including anti-bacterial and anti-fungal ac- showed that the ScBx genes are similar to those of other tivity, and the ability to reduce alfa–amylase activity. The bio- Poaceae species, especially to the TaBx genes. The polymor- synthesis and modification of BX are controlled by the genes phisms present both in the coding sequences and non-coding Bx1 ÷ Bx10, GT and glu, and the majority of these Bx genes regions of ScBx in relation to other Bx genes are predicted to have been mapped in maize, wheat and rye. However, the have an impact on the expression, structure and properties of genetic basis of BX biosynthesis remains largely the encoded proteins. uncharacterized apart from some data from maize and wheat. -

The Genetic Background of Benzoxazinoid Biosynthesis in Cereals
Acta Physiol Plant (2015) 37:176 DOI 10.1007/s11738-015-1927-3 REVIEW The genetic background of benzoxazinoid biosynthesis in cereals 1 1 1 B. Makowska • B. Bakera • M. Rakoczy-Trojanowska Received: 8 July 2014 / Revised: 11 May 2015 / Accepted: 22 July 2015 / Published online: 6 August 2015 Ó The Author(s) 2015. This article is published with open access at Springerlink.com Abstract Benzoxazinoids (BXs) are important com- Occurrence, characterization and biological role pounds in plant defense. Their allelopathic, nematode of BXs suppressive and antimicrobial properties are well known. BXs are found in monocot plants and in a few Benzoxazinoids (BXs) are protective and allelophatic sec- species of dicots. Over 50 years of study have led to ondary metabolites found in numerous species belonging to the characterization of the chromosomal locations and the Poaceae family, including maize, rye, wheat (Niemeyer coding sequences of almost all the genes involved in 1988a;Gru¨netal.2005;Nomuraetal.2007;Freyetal. BX biosynthesis in a number of cereal species: 2009; Chu et al. 2011; Sue et al. 2011), the wheat progen- ZmBx1–ZmBx10a7c in maize, TaBx1–TaBx5, TaGT itors Triticum urartu, Aegilops speltoides, Aegilops squar- and Taglu in wheat, ScBx17ScBx5, ScBx6-like, ScGT rosa (Niemeyer 1988b), and wild barleys: Hordeum and Scglu in rye. So far, the ortholog of the maize Bx7 roshevitzii, Hordeum flexuosum, Hordeum brachyantherum, gene has not been identified in the other investigated Hordeum lechleri (Gru¨netal.2005) and in genera—Chus- species. This review aims to summarize the available quea, Elymus, Arudo (Zu`n˜iga et al. 1983), Coix (Nagao et al. -

The Crop Trust 2015
THE CROP TRUST ANNUAL REPORT 2015 SUMMARY WWW.CROPTRUST.ORG/2015 1 THE CROP TRUST ANNUAL REPORT 2015 DEAR FRIENDS In this historic year, the nations of the world committed to a guiding set of Sustainable Development Goals and to action against the challenges of climate change. And the Crop Trust continued in its commitment to safeguard the agrobiodiversity that lies at the very root of sustainable development and climate adaptation alike. What did that commitment look like in 2015? The sound administrative and financial management of our partner genebanks got sounder. The Crop Wild Relatives project reached its peak with a full program of action across some 30 countries. Genesys, the global portal to information about crop collections, got more powerful and easier to use. The Svalbard Global Seed Vault saw new deposits, and its first retrieval – of important material from the genebank of the International Center for Agricultural Research in the Dry Areas (ICARDA) in Aleppo, Syria. Seeing those seeds sprout again will be an occasion for joy. There are solutions to our world’s challenges to be found in the diversity of life – as long as we keep it alive. With thanks to all our supporters, MARIE HAGA AND WALTER FUST 2 From the farmers of the Sacred Valley of Peru to The Prince of Wales, in 2015 our partners left us with no doubt that the work we do, and the ‘‘ global system we are building together, is all at once inspiring, exciting, and absolutely essential. MARIE HAGA, EXECUTIVE DIRECTOR ,, This year we continued to build up our support to crop collections, their inter-linkage in a global system for ex situ conservation, and the ‘‘ accessibility of all the diversity they hold. -

Policy Brief Global Conservation Priorities for Crop Wild Relatives
Policy Brief Global Conservation Priorities for Crop Wild Relatives Key Facts and Recommendations: • First of its kind global mapping effort reveals over 70% of essential crop wild relative species are under- represented in genebanks worldwide leaving food security unnecessarily vulnerable to future climate change • Collection efforts to preserve wild species with resilient traits useful for improving our crops ability to tolerate heat and pests must be stepped up • Long-term global food security requires inter- national collaboration on collecting, conserva- tion, and sharing these valuable genetic resources. • Countries are invited to use findings as a baseline to measure their progress on development and conservation commitments e.g. target 2.5 of the UN Sustainable Development Goals Many of the wild plant species at the foundation of the global food supply are missing from the world’s genebanks, according to new research by the International Center for Tropical Agriculture (CIAT) in coordination with the Global Crop Diversity Trust (Crop Trust) and the Royal Botanic Gardens (RBG), Kew. Crop wild relatives (CWR) are distant cousins of well-known food crops like rice, potato and wheat that contain traits such as drought and heat tolerance, and pest and disease resistance. This genetic diversity makes them an invaluable resource for plant breeders working to develop crops capable of adapting to the impacts of climate change, including higher temperatures, increased soil salinity due to rising sea levels, and more frequent and severe pest and disease outbreaks. The inability to access these crop wild relatives removes options to improve crops, leaving our food supply, and agricultural producers, vulnerable. -

Crop Trust Briefing
Agenda Item 12 For Information Re-Issued: 08 May 2018 Future Funding of CGIAR Genebanks: Background Document-Revision 1 Purpose This revised paper sets out the long-term fundraising goal of the Crop Trust, the commitments made to the Genebank Platform to 2022 and the short-term challenges to meet the fundraising target of $500 million by 2022 in order to fund the Article 15 collections in perpetuity. Action requested The System Council is asked to consider this background information ahead of the Crop Trust’s presentation to the System Council’s 6th meeting on the topic “Future Funding of CGIAR Genebanks”. Document category: Working document of the System Council There is no restriction on the circulation of this document Prepared by: Crop Trust 6th CGIAR System Council Meeting Revision1-SC6-08 16-17 May 2018, Berlin, Germany Page 1 of 16 Future Funding of CGIAR Genebanks Background Document - System Council 17 May 2018 Meeting Prepared by the Global Crop Diversity Trust 7 May 2018 1. Purpose The purpose of this paper is to provide System Council Members with background information ahead of the Crop Trust’s presentation to Members on 17 May 2018 on the topic “Future Funding of CGIAR Genebanks”. This paper sets out the long term fundraising goal of the Crop Trust, the commitments made to the Genebank Platform to 2022 and the short-term challenges to meet the fundraising target of $500 million by 2022 in order to fund the Article 15 collections under the International Treaty of Plant Genetic Resource (ITPGR) in perpetuity. 2. Fundraising Targets The Crop Trust’s Strategic Workplan sets out a target of USD 850 million required in the endowment fund in order to fund the operation of a global portfolio of national and international collections for the ex-situ conservation of crop diversity. -

Large Trees, Supertrees and the Grass Phylogeny
LARGE TREES, SUPERTREES AND THE GRASS PHYLOGENY Thesis submitted to the University of Dublin, Trinity College for the Degree of Doctor of Philosophy (Ph.D.) by Nicolas Salamin Department of Botany University of Dublin, Trinity College 2002 Research conducted under the supervision of Dr. Trevor R. Hodkinson Department of Botany, University of Dublin, Trinity College Dr. Vincent Savolainen Jodrell Laboratory, Molecular Systematics Section, Royal Botanic Gardens, Kew, London DECLARATION I thereby certify that this thesis has not been submitted as an exercise for a degree at any other University. This thesis contains research based on my own work, except where otherwise stated. I grant full permission to the Library of Trinity College to lend or copy this thesis upon request. SIGNED: ACKNOWLEDGMENTS I wish to thank Trevor Hodkinson and Vincent Savolainen for all the encouragement they gave me during the last three years. They provided very useful advice on scientific papers, presentation lectures and all aspects of the supervision of this thesis. It has been a great experience to work in Ireland, and I am especially grateful to Trevor for the warm welcome and all the help he gave me, at work or outside work, since the beginning of this Ph.D. in the Botany Department. I will always remember his patience and kindness to me at this time. I am also grateful to Vincent for his help and warm welcome during the different periods of time I stayed in London, but especially for all he did for me since my B.Sc. at the University of Lausanne. I wish also to thank Prof. -

Variation of Nitrate Reductase Genes in Selected Grass Species
Variation of nitrate reductase genes in selected grass species Jizhong Zhou, Andrzej Kilian, Robert L. Warner, and Andris Kleinhofs Abstract: In order to study the variation of nitrate reductase (NR) genes among grass species, gene number, intron size and number, and the heme-hinge fragment sequence of 25 grass species were compared. Genomic DNA cut with six restriction enzymes and hybridized with the barley NAD(P)H and NADH NR gene probes revealed a single NAD(P)H NR gene copy and two or more NADH NR gene copies per haploid genome in most of the species examined. Major exceptions were Hordeum vulgare, H. vulgare ssp. spontaneum, and Avena strigosa, which appeared to have a single NADH NR gene copy. The NADH NR gene intron number and lengths were examined by polymerase chain reaction amplification. Introns I and I11 appeared to be absent in at least one of the NADH NR genes in the grass species, while intron I1 varied from 0.8 to 2.4 kilobases in length. The NADH NR gene heme-hinge regions were amplified and sequenced. The estimated average overall nucleotide substitution rate in the sequenced region was 7.8 X lo-'' substitutions/site per year. The synonymous substitution rate was 2.11 X substitutions/synonymous site per year and the nonsynonymous substitution rate was 4.10 X lo-'' substitutions/nonsynonymous site per year. Phylogenetic analyses showed that all of the wild Hordeum species examined clustered in a group separate from H. vulgare and H. vulgare ssp. spontaneum. Key words: nitrate reductase gene, gene copy number, intron, molecular phylogeny, grasses.